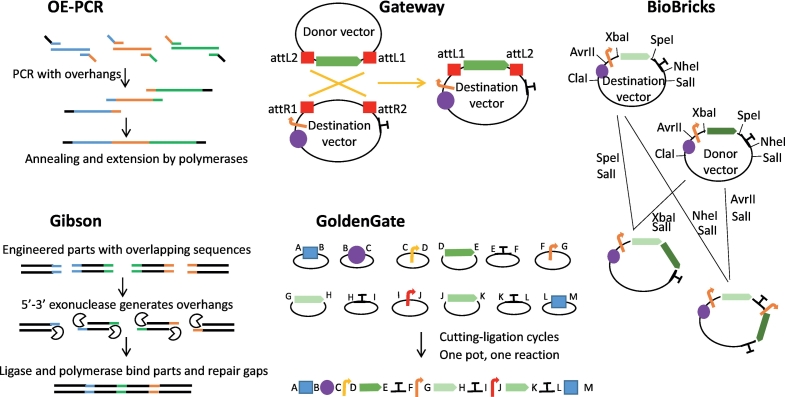
Fig. 1.
Summary of DNA assembly techniques.
OE-PCR is a two-step PCR. During the first step, complementary overlapping overhangs are added to the parts to be assembled. During the second step, the parts hybridize with each other and form the new assembly via extension. In the Gateway method, the gene of interest, which has been cloned into the entry vector, is transferred into the destination vector via att site recombination. The expression vector obtained is then digested to release the expression cassette and used to transform Y. lipolytica. The BioBricks technique is used to clone parts via restriction digestion and the subsequent ligation of the resultant compatible sticky ends. YaliBricks vectors were designed to have AvrII, XbaI, SpeI, and NheI endonuclease site recognition. The ligation of the compatible overhangs produces a scar that is no longer recognized by either enzyme, which allows for subsequent assembly steps using more DNA parts. In Gibson assembly, parts are synthesized to overlap by 30+ bp. Their ends are then processed by an exonuclease that creates single-stranded 3′ overhangs, which facilitates annealing. The overhangs are fused together using a polymerase, which fills in gaps within each annealed fragment; a ligase seals gaps in the assembled DNA. Golden Gate assembly exploits type II enzymes, which cut outside their recognition sites to excise parts with arbitrarily defined four-base overhangs. Through the careful selection of compatible overhangs, such parts can be assembled altogether in a defined order. In the figure, the letters A to M represent different compatible 4-nt overhangs; the yellow, orange, and red arrows represent promoters; the green arrows represent genes; the violet circles represent markers; the blue squares represent insertion sequences; and the Ts represent terminators.