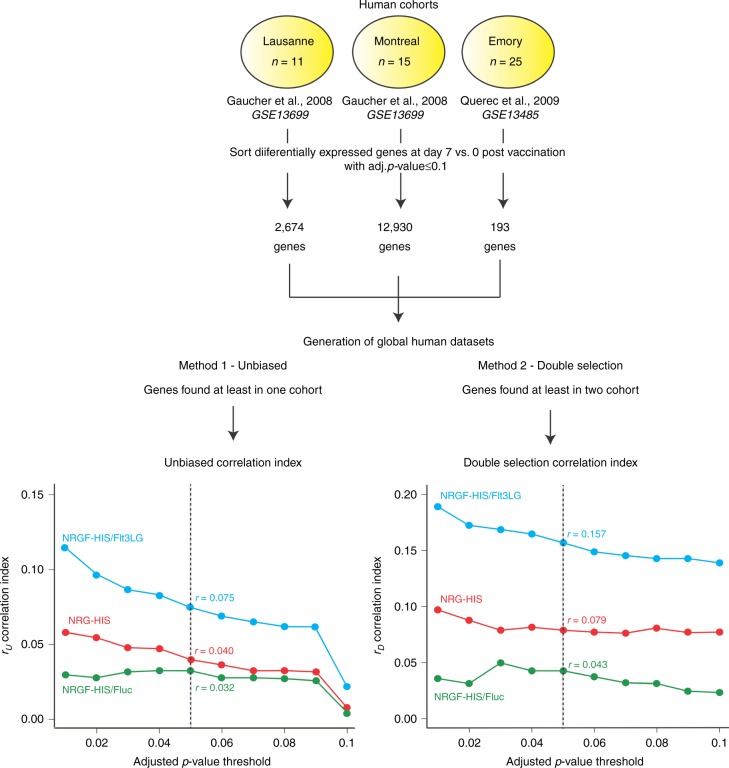
Fig. 5.
Human-like transcriptomic response to YFV-17D infection in NRGF-HIS/Flt3LG. Differentially expressed genes from PBMCs of three YFV-17D human vaccinee cohorts (Lausanne, n = 11; Montreal, n = 15; Emory, n = 25) were used to generate two global human transcriptomic datasets: an unbiased dataset (all differentially expressed genes (padj ≤ 0.1) found in at least one cohort) and an double-selection dataset (all differentially expressed genes (padj ≤ 0.1) found in at least two cohorts). For a given human global dataset (unbiased, left; double selection, right) and padj threshold (starting at padj ≤ 0.1 with 0.01 increment), the Spearman rho correlation index was computed for each humanized mouse model transcriptomic dataset (NRG-HIS, NRGF-HIS/Fluc, and NRGF-HIS/Flt3LG) and plotted as function of the padj threshold. The Spearman rho correlation index for the unbiased and double selection method are respectively referred as rU and rD. The definitive correlation indexes for the unbiased and double selection method, referred as rU and rD at padj = 0.05 (rU,q=0.05 and rD,q=0.05) respectively, are indicated on each graph and for each humanized mouse model. See Methods for more details