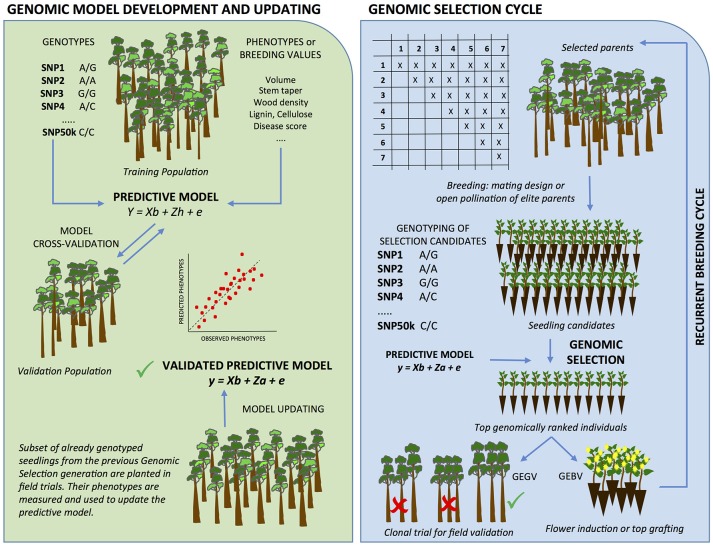
Figure 1.
Genomic selection in forest trees. GS begins with the development of a predictive model for the traits of interest (Left panel), which are then used in the GS cycles (Right panel) and progressively updated. GS uses genome-wide markers whose effects on the phenotype are estimated concurrently in a large and representative “training population” of individuals without applying severe significance tests. Markers are retained as forecasters of phenotypes in prediction models to be later applied to “selection candidates” for which only genotypes are collected. The prediction models are cross-validated against a “validation population,” a set of individuals of the same reference population that were not used for the estimation of marker effects. Once a prediction model is shown to provide adequate accuracy, it can be used in the GS cycle. An array of selection candidates - full of half-sib families derived from crossing either the original elite parents of the training set, or elite individuals selected in the training set - are genotyped and have their breeding values (GEBV) and/or genotypic values (GEGV; additive + non-additive effects) estimated using the model developed earlier. Top ranked seedlings for GEBV are subject to early flower induction and inter-mated to create the next generation of breeding. Top ranked seedlings for GEGV are clonally propagated and tested in verification clonal trials where elite clones are eventually selected for operational plantation. Additionally, all or subsets of the already genotyped selection candidates are planted in experimental design and phenotyped at the target selection age to provide genotype and trait data for GS model updating as GS generations advance and climate changes.