FIGURE 5.
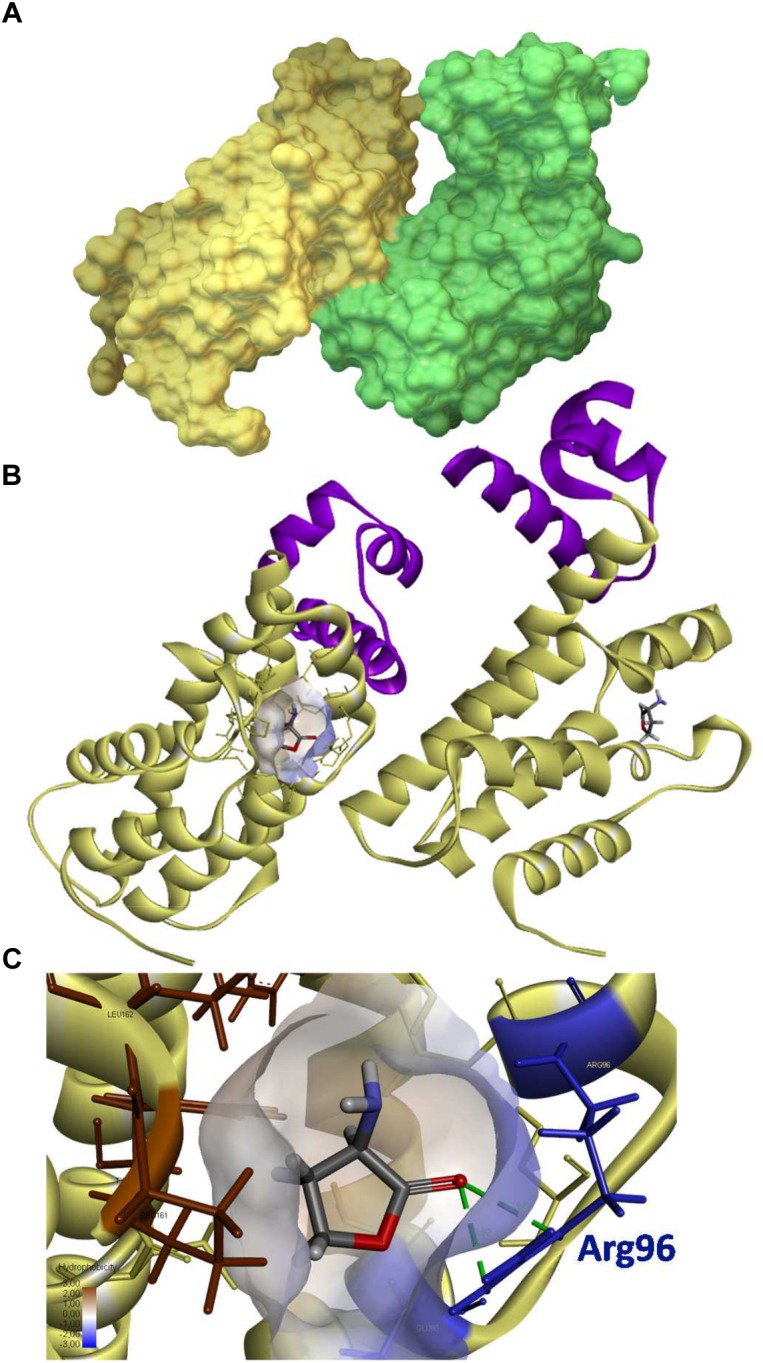
3D structure of QsdR and its potential ligand. The QsdR structure of the R138 strain (RCSB PDB accession number 4ZA6) was elucidated after crystallization and X-Ray diffraction (El Sahili et al., 2015). (A) A solvent-accessible surface area representation shows the functional structure of QsdR consisting of two monomers (yellow and green) that interact asymmetrically. (B) A secondary structure representation shows that each monomer presents both a DNA-binding domain (N-domain) colored in purple and an effector binding domain (C-domain) indicated in yellow, this last one harboring at the core a narrow half apolar/half polar cavity (drawn only on one of the monomers). (C) A close-up of the core cavity and HSL ligand are presented. Molecular docking methodology predicts a binding of the homoserine lactone (HSL), which is also the cyclic moiety of the QS signal, in each of the effector pockets (B,C). The lactone ester bond of the homoserine lactone is oriented toward the hydrophilic part of the pocket (colored in translucent blue) while the rest of the ligand molecule is oriented toward the hydrophobic part (colored in translucent brown). The hydrogen bonds between the oxygen atom of lactone carbonyl and the nitrogen atoms of Arg96 residue are indicated in green dashed line.
