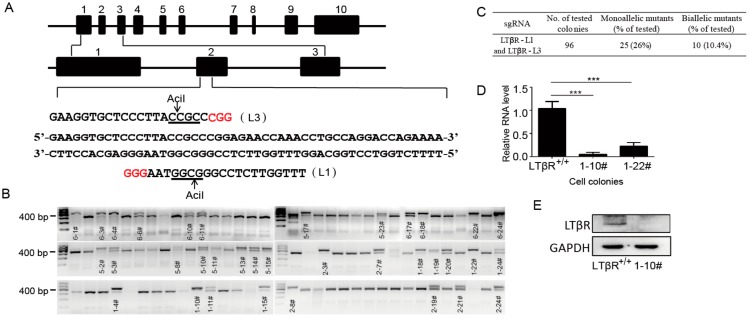
Figure 2.
Generation and identification of LTβR knockout intestinal porcine enterocyte (IPEC-J2) cells. (A) Targeting strategy: two different small guide RNAs (sgRNAs) (L1 and L3) targeting 32 bp regions of exon 2 in the pig LTβR gene were designed and inserted into the pX330 vector. Then, the pCAG-GFP plasmid was co-transfected with the pX330-L1 and pX330-L3 plasmids, and single cells were sorted into 96-well plates by flow cytometry. (B) A total of 96 colonies were picked and subjected to RFLP analysis. Monoallelic and biallelic mutant clones were labeled by their clone number. (C) Estimation of targeting efficiency. Note that 10 biallelic mutated clones were identified, and the targeting efficiency was 10.4%. (D) Wild-type IPEC-J2 cells and two biallelic mutated clones (namely 1-10# and 1-22#) were picked for real-time PCR analysis. *** p < 0.001 was considered statistically significant. Note that the expression levels of LTβR were significantly decreased in 1-10# and 1-22# cells (p < 0.001 respectively). (E) LTβR protein levels in the wild-type and 1-10# cells were detected by immunoblotting with an anti-LTβR antibody at a 1:1000 dilution (Abcam). GAPDH was used as a loading control.