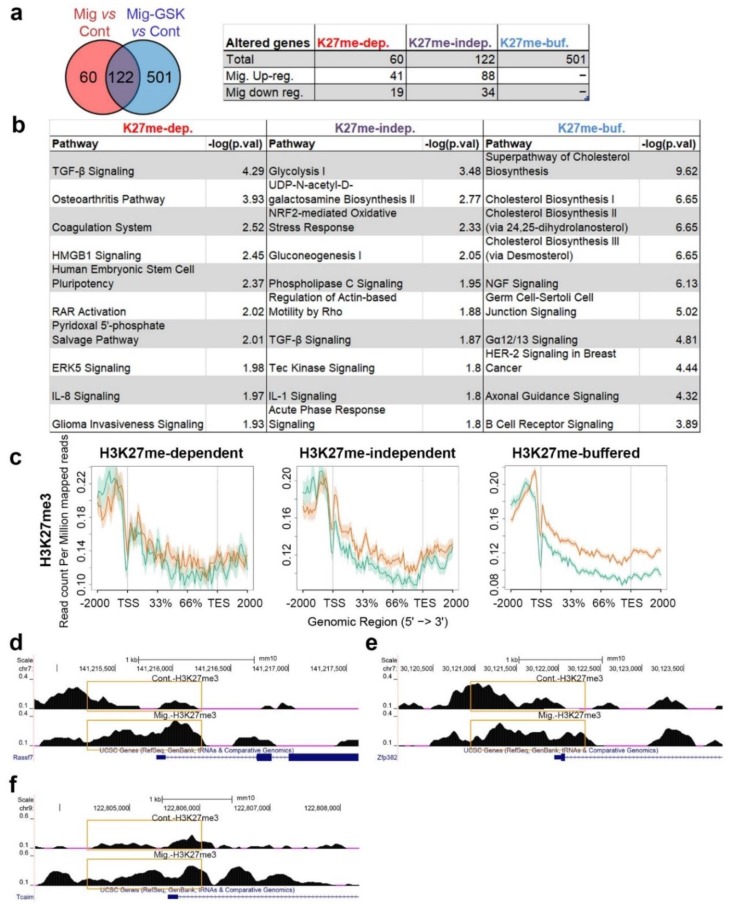
Figure 5.
The dependence of migration-induced transcriptional changes on H3K27 methylation. (a) Altered genes upon induction of migration (fold change > 1.3, FDR < 0.05) that were detected in untreated migrating cells and in migrating cells treated with GSK343 are represented by a Venn diagram. Overlapping genes are termed “H3K27me-independent”, genes altered only when migrating cells were compared to control cells are termed “H3K27me-dependent”, and differentially expressed genes only in GSK343-treated migrating cells are termed “H3K27me-buffered”. (b) Top 10 pathways altered according to the “H3K27me-independent” gene list, the “H3K27me-dependent” gene list, and the “H3K27me-buffered” gene list as identified by IPA. (c) Distribution of H3K27me3 signal across “H3K27me-dependent”, “H3K27me-independent” and “H3K27me-buffered” genes in control cells (green) and in migrating cells (orange). The region between 5’End and the 3’End represents genes bodies. Additional 2000 bp upstream and downstream of the genes are plotted. (d–f) H3K27me3 signal around the transcription start site (TSS) of Rassf7, Zfp382, and Tcaim. The altered signals around the TSS are marked by orange rectangles.