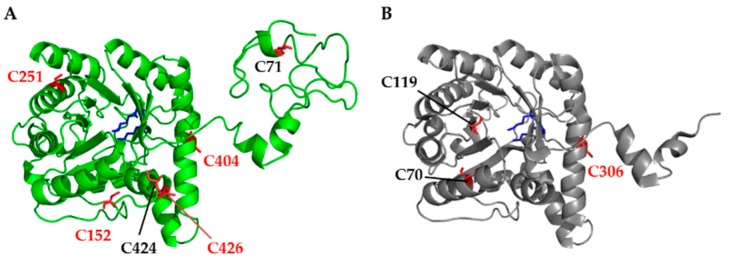
Figure 1.
Comparative analysis of the X-ray 3D structure of Chlorobium vibrioforme 5-aminolevulinic acid dehydratase (ALAD) and the modelled ALAD from Arabidopsis thaliana. (A) The structure of mature Arabidopis ALAD was visualized with PyMOL (Schrödinger) after structure prediction with Phyre2. The modeled structure shows high structural homology with the two X-ray structures of the Mg2+ dependent ALADs from Pseudomonas aeruginosa [32,33] and Chlorobium vibrioforme [34,35]. (B) The Chlorobium vibrioforme ALAD structure based on RCSB protein data bank (PDB) entry 1W1Z [34]. Cysteines are highlighted by red sticks, conserved cysteines in higher plants are highlighted in red lettering. The two Schiff base lysine residues (for A. thaliana K298 and K351) are highlighted in the two structures by blue sticks.