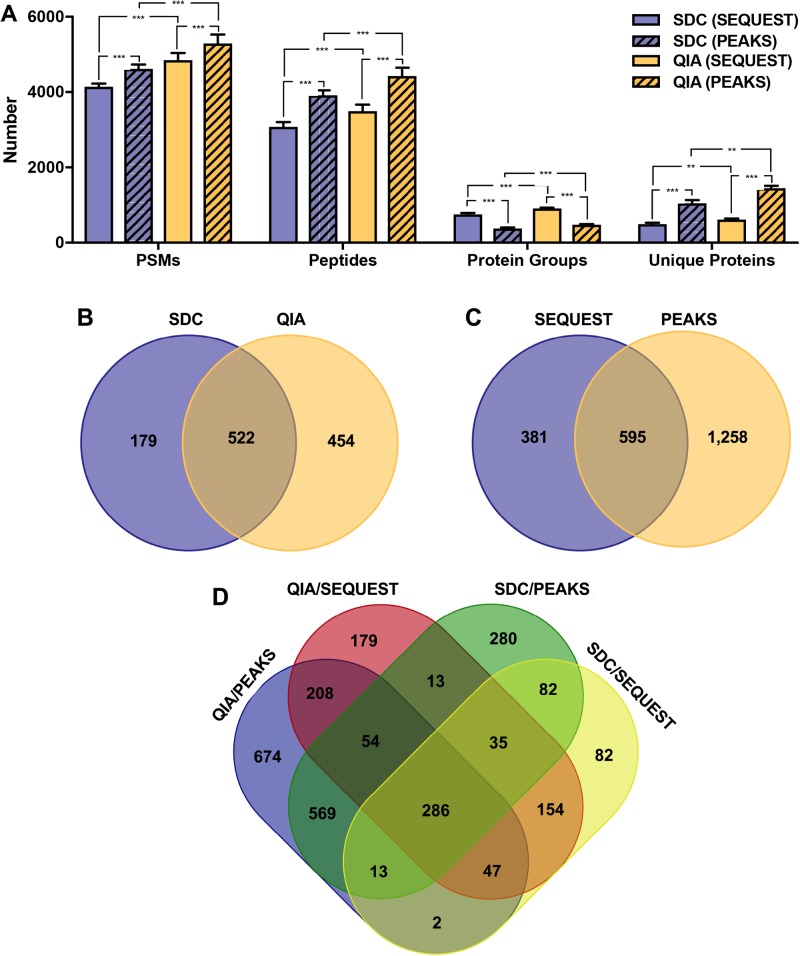
Fig. 2.
Comparison of proteins identified using two sample preparation and two data analysis workflows. (A) Numbers of peptide spectrum matches (PSMs) and peptides, protein groups, and unique proteins (with two or more unique peptide hits) identified by SEQUEST or PEAKS in samples prepared using SDC and QIA. SwissProt (2/13/2018) database was used for searches and only hits with false discovery rates (FDR) <1% were retained. Asterisks denote significant differences between sample preparation methods and software platforms: ***P<0.0001, **P<0.001. Sets of identified proteins were compared between (B) SDC and QIA methods and SEQUEST database search, (C) SEQUEST and PEAKS protein identification methods for QIA samples, and (D) between each sample preparation method and software platform.