Fig. 2. Establishment of latently infected T98G cells with rHCMV (T98G-LrV).
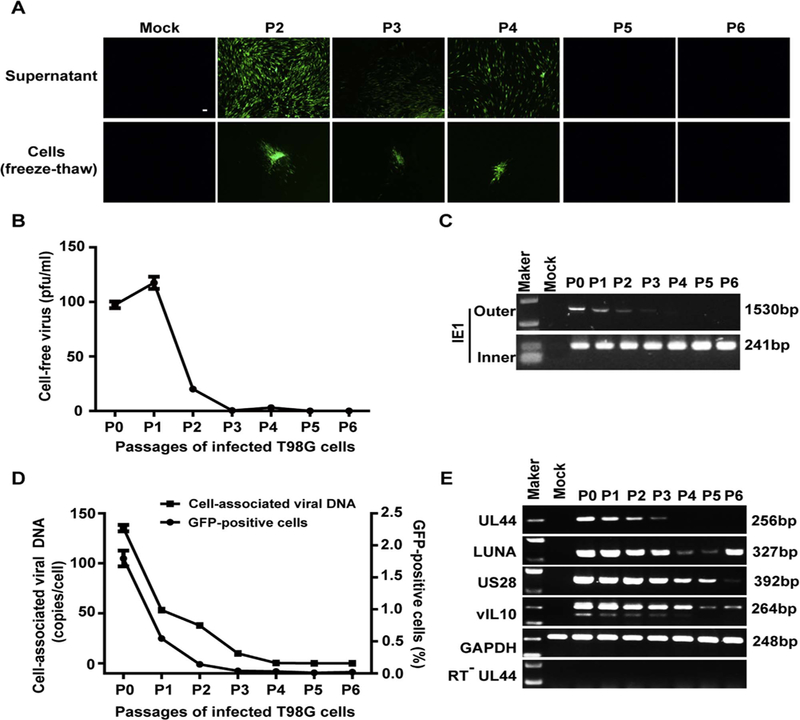
T98G cells were infected with rHCMV at an MOI of 10 and passaged every 7 days. Supernatants and cells of each passage were harvested before subculture. (A) Infectious virus. HFFs monolayers were inoculated with the supernatants (upper panel) or the infected cells (frozen-thawed cells, lower panel). GFP-positive foci were observed 14 days post inoculation. Scale bar: 100 µm. (B) Cell-free virus titer. Virus titers in the supernatants were determined by plaque forming assay. (C) Viral DNA. Cellular DNA was extracted from the infected cells of each passage, and viral genomic DNA was examined by nested PCR based on IE1 gene. Shown are the representative results of the first (Outer) and nested (Inner) amplification of PCR, respectively. (D) Cell-associated viral DNA and GFP-positive cells. Viral genome copy numbers in the infected cells were determined by qPCR. Serial dilution of T98G cells was isolated for DNA and subjected to qPCR for determination the GAPDH copy number of each cell. Viral genome copies were calculated as “copies/cell” with adjusting to GAPDH. GFP-positive cells at each passage was analyzed by FACS. (E) Transcription of representative viral genes. Total RNA was extracted from each set of the passaged cells, reversely transcribed to cDNA, and assayed for the indicated genes by PCR. The mRNA without reverse transcription (RT−) were assayed for UL44 as control to exclude viral DNA contamination.
