Abstract
In the last few decades, a large body of experimental evidence has highlighted the complex role for mitochondria in eukaryotic cells: they are not only the site of aerobic metabolism (thus providing most of the ATP supply for endergonic processes) but also a crucial checkpoint of cell death processes (both necrosis and apoptosis) and autophagy. For this purpose, mitochondria must receive and decode the wide variety of physiological and pathological stimuli impacting on the cell. The “old” notion that mitochondria possess a sophisticated machinery for accumulating and releasing Ca 2+, the most common and versatile second messenger of eukaryotic cells, is thus no surprise. What may be surprising is that the identification of the molecules involved in mitochondrial Ca 2+ transport occurred only in the last decade for both the influx (the mitochondrial Ca 2+ uniporter, MCU) and the efflux (the sodium calcium exchanger, NCX) pathways. In this review, we will focus on the description of the amazing molecular complexity of the MCU complex, highlighting the numerous functional implications of the tissue-specific expression of the variants of the channel pore components (MCU/MCUb) and of the associated proteins (MICU 1, 2, and 3, EMRE, and MCUR1).
Keywords: Mitochondria, mitochondrial Ca2+ uptake, mitochondrial ion transport, MCU, MICU
Introduction
Ca 2+ is universally recognised as one of the most pleiotropic second messengers in cell biology. Indeed, Ca 2+ ions are responsible for decoding a variety of extracellular and intracellular stimuli, which, in animals, range from endocrine secretion to gene expression, muscle contraction, and synaptic transmission 1– 5. The efficacy of Ca 2+ as a signalling molecule relies mainly on the maintenance of a steep Ca 2+ gradient between the concentration in the extracellular (few mM) and intracellular (∼100 nM) environments. This >10,000-fold difference ensures that even very small changes in intracellular Ca 2+ concentration ([Ca 2+] i) are effective in regulating the numerous Ca 2+-sensitive proteins of the cell, such as catalytic enzymes, channels, and transcription factors 6. The maintenance of a low [Ca 2+] i is tightly controlled by the presence of pumps and transporters both at the plasma membrane 7 and at the membrane of organelles accumulating large amounts of Ca 2+ 8, 9. The endoplasmic/sarcoplasmic reticulum (ER or SR in striated muscle) is undoubtedly the main intracellular Ca 2+ store; nevertheless, other cellular compartments actively participate in modulating [Ca 2+] i: first of all mitochondria but also the Golgi apparatus 10, endosomes, and lysosomes 2, 5, 11.
The role for mitochondria in Ca 2+ handling was first demonstrated in the 1960’s, when their ability to actively accumulate Ca 2+ was evaluated in different cellular ex vivo models 12– 14. This occurred even before the advent of the chemiosmotic theory 15, which provided the thermodynamic basis for Ca 2+ entry into mitochondria. In addition, the role for mitochondria in cell function has gradually expanded, and its initial identification as the powerhouse for cellular energy supply was subsequently integrated into a more complex activity that includes the regulation of cell death, metabolism, and signalling pathways 16– 18. Notably, many mitochondrial functions are directly regulated by the level of Ca 2+ ions inside the organelles 4, 19. The control of mitochondrial Ca 2+ concentration ([Ca 2+] mt) is thus of primary relevance for cell physiology, and emerging evidence converges on the concept that its dysregulation is of utmost importance in the establishment of pathological conditions.
In this commentary, we will focus on the molecular machinery underlying mitochondrial Ca 2+ uptake, the mitochondrial Ca 2+ uniporter (MCU) complex (see Figure 1), to which our laboratory has dedicated much effort in the last decade, contributing to its discovery in 2011 and unravelling its molecular complexity and functional significance.
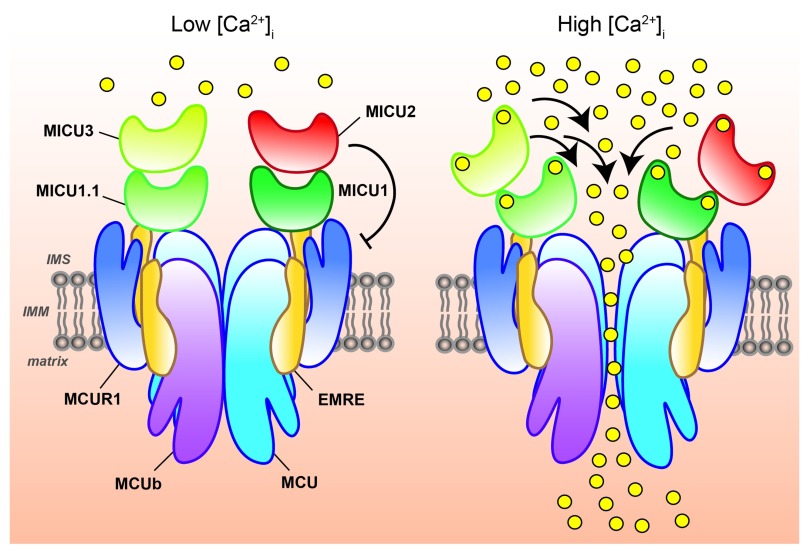
Figure 1. The mitochondrial calcium uniporter (MCU) complex.
Schematic representation of MCU-mediated Ca 2+ entry into mitochondria at different intracellular Ca 2+ concentrations ([Ca 2+] i). Mitochondrial Ca 2+ uptake is controlled by a multiprotein complex consisting of MCU and MCUb (the pore-forming subunits) together with the essential mitochondrial Ca 2+ uniporter regulator (EMRE), the mitochondrial Ca 2+ uptake (MICU) proteins, MICU1, MICU1.1, MICU2, and MICU3, and, possibly, the MCU regulator 1 (MCUR1). At low [Ca 2+] i, MICU1/MICU1.1–MICU2 or MICU1/MICU1.1–MICU3 heterodimers ensure MCU gatekeeper activity, preventing undesirable mitochondrial Ca 2+ cycling in resting cells. At high [Ca 2+] i, the MICU proteins act as positive regulators of MCU channel activity, allowing efficient mitochondrial Ca 2+ uptake (right). IMS, intermembrane space; IMM, inner mitochondrial membrane.
The mitochondrial Ca 2+ uniporter complex
Since the first reports of mitochondrial Ca 2+ uptake 12– 14, extensive studies have been conducted to identify the mitochondrial Ca 2+ entry mechanism, characterised by Ruthenium Red sensitivity, high selectivity, and high capacity for the cation 20. Only from 2010, with the identification of the first regulator of the mitochondrial Ca 2+ channel, the mitochondrial Ca 2+ uptake protein 1 (MICU1) 21, and, one year after, by the cloning and molecular characterisation of the gene coding for the MCU 22, 23, the entry pathway was characterised at the molecular level and the route for the genetic manipulation of mitochondrial Ca 2+ uptake was finally open.
The mitochondrial Ca 2+ uniporter MCU
In 2011, two independent in silico screenings 22, 23 identified the CCDC109a gene product as the long-sought pore-forming unit of the MCU channel mediating mitochondrial Ca 2+ uptake in mammalian cells. Purified MCU protein reconstitution in lipid bilayers revealed a Ca 2+ channel activity with the electrophysiological features of the hypothetical MCU 20. In addition, it was clearly shown that MCU downregulation in mammalian cells inhibits mitochondrial Ca 2+ uptake while its overexpression increases mitochondrial Ca 2+ accumulation, thus proving that it is bona fide the channel mediating Ca 2+ uptake in mitochondria 23.
The sequence of MCU consists of two transmembrane α-helix domains spanning the inner mitochondrial membrane (IMM), the second of which contains the critical DIME motif responsible for the channel selectivity filter, linked by a short loop toward the mitochondrial intermembrane space (IMS), while both the C- and the N-termini face the mitochondrial matrix 22, 24. A single MCU molecule per se is thus not able to form the channel but should organise in oligomers, and MCU was initially proposed to arrange itself into tetramers 25. This hypothesis was very recently confirmed by coherent high-resolution cryo-EM data from four independent groups on different MCU orthologs 24, 26– 28. These seminal studies finally unveiled the structure and arrangement of MCU protomers within the complex and the exact position of the channel selectivity filter. The 3D reconstruction of EM data unambiguously revealed that MCU forms tetramers with a nonobvious symmetry: the transmembrane domain displays a fourfold symmetry, while the N-terminal domain (NTD) on the matrix side shows a twofold symmetry axis. These findings confuted the pentameric MCU architecture that was previously proposed based on magnetic resonance and negative staining EM data from Caenorhabditis elegans MCU-ΔNTD protein, in which a significant portion of the NTD was removed to facilitate the analysis 29. In addition, the MCU DIME motif, which contains the two critical acidic residues directly involved in the cation coordination, has now been shown to reside at the beginning of––and thus integral to––the second transmembrane helix (TMH), and not in the loop connecting the two transmembrane helices, as previously suggested 29.
However, as Raffaello et al. showed, MCU is not the only pore-forming unit of the oligomer, but it associates with the protein encoded by the MCU paralog gene CCDC109b, which was thus named MCUb 25. The characterisation of MCUb demonstrated that it acts as a negative regulator of MCU activity both in lipid bilayer experiments and when overexpressed in mammalian cells 25 (see Figure 1). To note, in other organisms, such as Trypanosoma cruzi, the ortholog of MCUb acts as a Ca 2+ conducting subunit and its overexpression enhances, rather than dampens, mitochondrial Ca 2+ uptake 30. Interestingly, in another trypanosomatid, Trypanosoma brucei, two additional MCU isoforms were identified, MCUc and MCUd, which are also endowed with Ca 2+ uniporter activity and are able to form heterotetrameric complexes with their homologues MCU and MCUb 31. This highlights the existence of significant species-specific differences in the function and distribution of MCU orthologues, despite their sequence conservation, and this should be taken into account when studying different organisms. Moreover, the expression and relative proportion of MCU and MCUb vary significantly among different tissues in mammals; thus, each cell type may have different variants of the uniporter owing to the specific composition and stoichiometry of MCU subunits. This accounts for tissue-specific variations of the capacity of mitochondria to take up Ca 2+ and perfectly fits with the electrophysiological recordings of MCU Ca 2+ currents in mitochondria from different mammalian tissues 32. In skeletal muscle, for example, the presence of a high MCU:MCUb ratio 25 matches the highest mitochondrial Ca 2+ conductance recorded in this tissue 32, whereas, in adult heart, the relatively elevated MCUb expression 25 results in a considerably low Ca 2+ current in cardiomyocyte mitochondria 32. In cardiac cells, in which ∼37% of the volume is occupied by mitochondria 33, this is crucial for preventing massive mitochondrial Ca 2+ accumulation that would potentially cause undesired buffering of the [Ca 2+] i transients required for contraction, futile cycling of Ca 2+ across the IMM, and, eventually, organelle Ca 2+ overload and apoptosis. The control of the MCU:MCUb proportion in the mitochondrial Ca 2+ channel pore domain is thus fundamental for the function and physiology of different tissues.
After the molecular identification of the MCU pore components and auxiliary factors, the path for the genetic manipulation of mitochondrial Ca 2+ uptake machinery in cells and animal models was finally opened. The first MCU –/– mouse was generated by Finkel’s group in 2013. It shows a relatively mild phenotype, and, despite the expected abrogation of mitochondrial Ca 2+ uptake, it develops normally and displays unaffected basal metabolism 34. However, the MCU –/– mouse shows increased plasma lactate levels after starvation and impaired exercise performance accompanied by a reduction in the activity of pyruvate dehydrogenase in skeletal muscle 34. These findings, together with the fact that MCU –/– mouse survival depends on the genetic background ( MCU deletion is embryonically lethal in a pure C57/BL6 background 35), pointed to a more subtle and less obvious involvement of mitochondrial Ca 2+ uptake in organ metabolism and organism development. This issue has been elegantly addressed by recent studies implementing tissue-specific modulation of MCU expression (by ablation/downregulation or overexpression) in adult tissues. For example, cardiac-specific tamoxifen-inducible MCU deletion in adult mice, differently from the germline genetic ablation of MCU, which did not protect from ischemic-reperfusion injury 34, clearly protects from ischemia-reperfusion heart damage, abrogates the contractile responsiveness to β-adrenergic stimulation responsible for the so-called “fight-or-flight” response, and reduces heart bioenergetics reserve capacity, even though it does not induce phenotypic abnormalities either in basal conditions or after cardiac overload and does not alter cardiomyocytes’ resting [Ca 2+] mt 36– 39. These results suggest that MCU may be dispensable for cardiac homeostasis in basal conditions, while it appears to play a major role in cardiac metabolic flexibility during acute stress. Similarly to what has been reported for the heart tissue, germline deletion of MCU did not protect from hypoxic/ischemic brain injury 40. However, brains from conditional neuronal-specific inducible MCU-deleted mice as well as primary cortical neurons silenced for MCU show a significant reduction of the hypoxic/ischemic damage and decreased cell death without impairment of neuronal mitochondria metabolism 41. Once more, these findings point to the existence of a strong drive for organs and tissues to respond to chronic MCU ablation by establishing adaptive metabolic shunts in order to cope with/bypass impaired mitochondrial Ca 2+ signalling. Such adaptation to mitochondria dysfunction has also been described in the cardiac-specific Tfam –/– mouse, the murine model for human cardiomyopathies with mtDNA depletion 42, in which the alteration in mitochondrial metabolism due to oxidative phosphorylation (OXPHOS) deficiency induces a secondary upregulation of MCU protein and a concomitant downregulation of NCLX transcription 43. The resulting increased mitochondrial Ca 2+ uptake and reduced Ca 2+ efflux in Tfam –/– cardiac mitochondria, although inducing an elevated and potentially damaging [Ca 2+] mt, appear somehow functional to ensure an efficient Ca 2+-dependent mitochondrial respiration that otherwise would be compromised by the pathology 41.
Overall, these findings lead us to the following consideration: experimental systems featuring long-term genetic manipulation of the mitochondrial Ca 2+ uptake machinery should be evaluated with caution, and the potential induction of unpredicted phenotypes, as a consequence of adaptive/maladaptive responses, should be taken into account. This is particularly relevant when in vivo models are considered.
In line with this, the acute but transient manipulation of MCU expression in skeletal muscle provided additional intriguing evidence of the wide-ranging influence of mitochondrial Ca 2+ signalling in the maintenance of organ and organism homeostasis. The decrease or enhancement of [Ca 2+] mt by AAV-mediated MCU silencing or overexpression, respectively, demonstrated that mitochondrial Ca 2+ uptake regulates myofibre trophism through the modulation of the activity of the insulin growth factor-1/Akt pathway and the transcription of the peroxisome proliferator-activated receptor gamma coactivator 1 alpha 4 gene ( PGC1α4). In particular, the overexpression of MCU in both adult and neonatal muscles induces significant fibre hypertrophy, which is not accompanied by alterations in mitochondrial membrane potential or other metabolic parameters (such as glycogen content and succinate dehydrogenase activity) 44. On the contrary, MCU downregulation leads to marked fibre atrophy as demonstrated by reduction of fibre size, inhibition of Akt activity, reduction of PGC1α4 transcripts, and impaired activation of the pyruvate dehydrogenase complex 44, in agreement with the data from the MCU –/– mice 34 (see Figure 2).
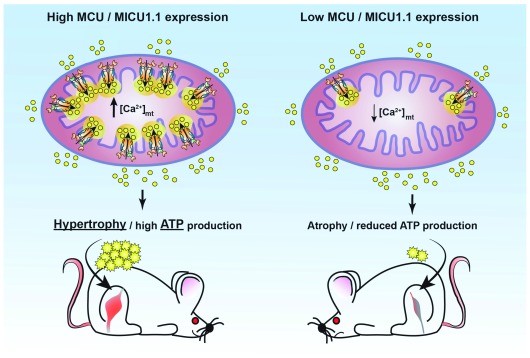
Figure 2. The role of mitochondrial Ca 2+ signalling in skeletal muscle trophism.
Schematic representation of the effects of manipulating mitochondrial Ca 2+ uptake in skeletal muscle fibres. Increased mitochondrial Ca 2+ accumulation by enhanced expression of mitochondrial Ca 2+ uniporter (MCU) or mitochondrial Ca 2+ uptake protein 1.1 (MICU1.1) leads to muscle hypertrophy by stimulating fibre growth and ATP production (left). Reduction of mitochondrial Ca 2+ uptake by downregulation of either MCU or MICU1.1 causes fibre atrophy and impaired ATP production (right). [Ca 2+] mt, mitochondrial Ca 2+ concentration.
Moreover, the recent description of a mouse model bearing constitutive skeletal muscle-specific deletion of MCU ( skMCU –/– mice) has been fundamental to uncover the metabolic rewiring occurring at the level of skeletal muscle cell but also, importantly, systemically 45. In line with previous data 34, 44, the muscle of skMCU –/– mice shows decreased performance and smaller fibre size compared to that of control mice 45. Moreover, it shows a shift toward fast fibre type and an enhanced preference for fatty acid oxidative metabolism 45. Interestingly, the ablation of MCU in skeletal muscle also impinges on other tissues, as shown by the increased catabolic response in both liver and adipose tissue of skMCU –/– animals 45. This clearly points out how the mitochondrial metabolic adaptation in response to altered Ca 2+ dynamics (i.e. MCU deletion) within the muscle tissue may have major systemic impact on different tissues and, importantly, on the modulation of global metabolism.
The MICU proteins
MICU1 was the first regulator of mitochondrial Ca 2+ uptake to be identified 21, even before the identification of MCU itself. It was initially described as an EF-hand Ca 2+-binding protein associated with the IMM that stimulates MCU channel opening and thus acts as a positive modulator of mitochondrial Ca 2+ uptake. Subsequent findings defined the crucial role of MICU1 in keeping the channel inactive under resting conditions, i.e. at low [Ca 2+] i 46, 47, thus preventing massive Ca 2+ entry that will otherwise cross the IMM down the steep electrochemical gradient and cause harmful mitochondrial Ca 2+ overload. MICU1 was thus appropriately defined as the gatekeeper of the MCU channel. This concept has been recently confirmed by two independent in vivo MICU1 –/– mouse models. These MICU1 –/– animals, despite normal embryonic development and growth, show a variable degree of postnatal lethality (one out of six to one out of seven surviving pups in the case of CRISPR/Cas9 mutants 48 and 100% lethality in the case of Cre recombinase/LoxP deleted alleles 49) mostly due to breathing failure for the underdevelopment of the respiration coordination centres (cerebellum and Purkinje cells) 49. The few surviving knockout mice display muscle impairment and ataxia 48, a phenotype very similar to that described in children with a MICU1 mutation 50. The analysis of MICU1 –/– mitochondria revealed increased matrix [Ca 2+] mt, increased sensitivity to permeability transition pore (PTP) opening and cell death, and reduced production of ATP, similar to what was reported in MICU1-silenced hepatocytes and Hela cells 46, 47. Overall, these data clearly highlight the relevance of MICU1 gatekeeping function in the control of mitochondrial Ca 2+ homeostasis and metabolism. Nevertheless, an additional intriguing feature of MICU1 action has recently emerged, which consists of its ability to confer MCU selectivity for Ca 2+ over Mn 2+. Indeed, the DIME motif in the MCU pore subunit appears to be unable per se to discriminate between Ca 2+ and Mn 2+ in the absence of MICU1 51. However, when present, MICU1 prevents Mn 2+ entry in mitochondria, thus ensuring the stringent Ca 2+ selectivity of MCU 51. In this scenario, the proper stoichiometry of MICU1 is thus crucial to avoid mitochondrial Mn 2+ uptake and to prevent Mn 2+ toxicity. This concept may have relevant implications for human pathologies, especially for those diseases caused by MICU1 deficiency 50, 52, 53 but also for Parkinson’s disease, as already suggested 51.
Additional MICU family members have been identified by genome sequence analysis soon after MICU1, namely MICU2 (EFHA1) and MICU3 (EFHA2) 54, which share 41% and 34% identity with MICU1, respectively, but display a broad and non-overlapping expression pattern compared to MICU1 55. In particular, MICU2 was shown to bind covalently to MICU1 through disulphide bridges, and the resulting dimer endows the genuine MCU gatekeeper activity 56. In this scenario, cytosolic Ca 2+ elevation causes Ca 2+ binding to the EF-hand domains of the MICU1–MICU2 dimer, allowing MICU1 to function as a positive regulator of MCU activity, thus efficiently promoting Ca 2+ entry into the organelle 56 (see Figure 1).
As for MICU3, its evolutionary conservation and sequence similarity to MICU1 suggested that it may share a common biological function with MICU1 54. Along this line, its role as a positive regulator of mitochondrial Ca 2+ uptake has been demonstrated recently 55. Indeed, the expression of MICU3 in Hela cells, which normally do not express it, showed that it interacts with MICU1, but not with MICU2, forming obligatory dimers 55 (see Figure 1). This interaction causes a significant increase of mitochondrial Ca 2+ uptake, demonstrating the stimulatory action of MICU3 on MCU activity. In addition, while displaying a reduced gatekeeper activity, MICU3 has been shown to mediate a more rapid response of mitochondria to [Ca 2+] i changes, allowing a shorter delay between the [Ca 2+] i rise and the increase in mitochondria Ca 2+ uptake compared to MICU1 55. MICU3 thus plays a pivotal role in determining the kinetics of mitochondrial Ca 2+ signalling and consequently in the modulation of global [Ca 2+] i dynamics, which may be of utmost relevance for the decoding of Ca 2+ signals in excitable cells, such as neurons, where indeed MICU3 shows the highest level of expression 54, 55.
By the interaction of monomers with non-overlapping functions, the MICU proteins confer an important property of the mitochondrial Ca 2+ uptake system: the sigmoidicity of the dose-response to Ca 2+. Indeed, the activity of MCU, which is kept low at resting [Ca 2+] i (∼100 nM) owing to the MICU1–MICU2 gatekeeper action, as already discussed, becomes extremely efficient upon [Ca 2+] i elevation. Indeed, Ca 2+ binding to the MICU1–MICU2 regulatory complex converts it from an inhibitory regulator (ensuring gatekeeping) at basal [Ca 2+] i to an activator, strongly enhancing the flux through the channel. The threshold for MCU activation is directly dependent on the Ca 2+ affinity of MICUs’ EF hands (which range from 300 nM to 1.3 µM), since the cooperative Ca 2+ binding to these domains induces a conformational change in the MICU1–MICU2 dimer, which acts as a molecular switch relieving MCU inhibition 57.
Despite the role for MICU2 in the regulation of MCU gatekeeping and activation being controversial still, an intriguing hypothesis is that it has evolved to spatially restrict the Ca 2+ crosstalk between single inositol trisphosphate receptor (IP3R) and MCU channels 58. Indeed, the presence of MICU2 has been shown to decrease the Ca 2+ affinity of the MICU1 gatekeeper, thus elevating the threshold for MCU opening 58. In the context of the intracellular environment, this increased threshold for mitochondrial Ca 2+ uptake translates into the need for a shorter distance between the ER Ca 2+-releasing channel (IP3R) and the mitochondrial Ca 2+ uptake channel (MCU) 58.
In line with this, in the last few years, the stoichiometry of MCU components has emerged as a crucial feature of the modulation of mitochondrial Ca 2+ uptake rate. The relative proportion of the MICU proteins, in particular, has been demonstrated as the basis of the tissue-specific differences in cellular Ca 2+ dynamics 59. Indeed, the different ratio of MICU1:MCU and MICU1:MICU2 proteins in the liver, the heart, and skeletal muscle has been correlated with the distinct mitochondrial Ca 2+ handling of these three tissues 59. The level of MICU1 expression, which is much lower in cardiac tissue (while MCU and MICU2 are present at similar levels) compared to liver, has been suggested to determine the lower maximal Ca 2+ uptake capacity and less-steep Ca 2+ dependence that characterise cardiac mitochondria compared to liver mitochondria, as electrophysiological data 32 and intracellular Ca 2+ measurements 59 revealed. The low amount of MICU1 in cardiac mitochondria is instrumental to reduce mitochondrial Ca 2+ uptake upon cytosolic Ca 2+ elevation, allowing the decoding of the repetitive cytosolic Ca 2+ spikes of the beating heart into a graded increase of matrix Ca 2+. This integration of frequency fluctuations is crucial to prevent mitochondrial Ca 2+ overload, which will be otherwise detrimental to the cardiac myocytes. By contrast, the relatively high MICU1 level and its cooperative activation of MCU in hepatocytes ensure the punctual and massive mitochondrial Ca 2+ increase at every single cytosolic Ca 2+ spike 59, which is functional to liver oxidative metabolism.
An additional degree of complexity came with the discovery of an alternative MICU1 isoform, extremely conserved and expressed at the highest level in skeletal muscle, which originates from the alternative splicing of the MICU1 mRNA, the MICU1.1 splice variant 60 (see Figure 1). This alternative isoform was shown to dimerise with MICU2, showing gatekeeper activity similar to that of MICU1, but it binds Ca 2+ one order of magnitude more efficiently than MICU1 and activates MCU at lower [Ca 2+] i than MICU1 60. Thus, in skeletal muscle, the MICU1.1–MICU2 dimer ensures a much higher net Ca 2+ entry and elevated ATP production than the MICU1–MICU2 dimer 60. These results highlight a novel mechanism of the molecular plasticity of the MCU Ca 2+ uptake machinery that allows skeletal muscle mitochondria to adapt to the intense metabolic challenge that characterises this tissue (see Figure 2).
The essential mitochondrial Ca 2+ uniporter regulator EMRE
To add further complexity to the study of MCU channel regulation, other molecules have been shown to interact with MCU in recent years. Indeed, the quantitative mass spectrometry analysis of the MCU-interacting proteins revealed the presence of a critical component of the MCU complex, the essential MCU regulator (EMRE) (see Figure 1), a 10 kDa single-pass transmembrane protein that was proposed to be necessary for MCU function by bridging MCU interaction with MICU1 in mammalian cells 61. Interestingly, EMRE appears to be required for MCU channel activity in metazoans only, since it is not found in plants, fungi, or protozoa, such as Dictyostelium discoideum, although they express MCU proteins with functional channel activity. While its exact role in the MCU complex is still uncertain, EMRE was proposed to mediate MCU sensitivity to matrix [Ca 2+] mt, suggesting that MCU activity may be modulated by Ca 2+ sensors facing both the IMS (MICUs) and the matrix (EMRE) 62. However, this hypothesis was subsequently questioned by other groups, whose data supported an opposite topology for the EMRE protein, with the short N-terminus exposed to the matrix while its acidic C-terminus faces the IMS 63, 64. In this conformation, EMRE action would be that of supporting MCU Ca 2+ transport activity by interacting with the first TMH of MCU within the IMM and to interact also with MICU1 at the IMS via its C-terminus poly-aspartate tail, thus ensuring MICU1 binding to the channel and gatekeeping activity 63.
Recently, the control of EMRE protein levels has emerged as a crucial aspect of MCU complex regulation. Indeed, a correct amount of EMRE is fundamental to guarantee the exact stoichiometry of the different MCU components. Alteration of EMRE protein levels by ablation of the AAA-proteases responsible for its degradation 65, or by expression of proteolytic-resistant EMRE constructs 62, leads to uncontrolled MCU channel opening causing mitochondrial Ca 2+ leakage and mitochondrial Ca 2+ overload, resulting in neuronal cell death 65, 66. The existence of a tight control of EMRE protein levels has also been demonstrated in vivo in the MICU1 –/– mice, where EMRE is reduced in a genetic condition characterised by increased [Ca 2+] mt basal levels 48. These findings clearly underline a negative regulatory mechanism that keeps EMRE expression in check in order to cope with changes in MCU activity 48.
The mitochondrial Ca 2+ uniporter regulator 1 MCUR1
Another protein that has been associated with the MCU complex 67, 68, despite the original proteomic analysis of the MCU interactors failed to recover it 61, is the MCU regulator 1 (MCUR1). MCUR1 is a coiled-coil-containing protein encoded by the CCDC90A gene initially identified by a RNAi screen searching for a mitochondrial membrane protein involved in MCU homeostasis 67, whose precise function in the regulation of MCU activity and mitochondrial metabolism is still debated. Indeed, MCUR1 has been shown to be required for MCU-dependent Ca 2+ uptake, since its knockdown dampens mitochondrial Ca 2+ entry while its overexpression enhances it 67. However, a direct action of MCUR1 on MCU activity was questioned by the finding that its silencing leads to a significant loss of mitochondrial membrane potential (ΔΨ m), which per se can account for the decreased mitochondrial Ca 2+ uptake 69. In this context, MCUR1 has been suggested to act primarily as a cytochrome c oxidase (COX) assembly factor 69, and this concept would also be supported by the fact that a MCUR1 orthologue is present in budding yeast 70, which lack MCU, and is required for yeast COX activity and survival in non-fermentable medium 69. Nevertheless, the patch clamp data of Ca 2+ currents from voltage-clamped mitoplasts point to an effective role for the MCUR1 protein on MCU channel conductance, independently of ΔΨ m 71. In line with this, experiments in Drosophila cells, which are resistant to Ca 2+-induced mitochondrial permeability transition (MTP) 72 and in which no MCUR1 homologs are found, show increased sensitivity to Ca 2+-dependent PTP opening when heterologous human MCUR1 is expressed, which is not accompanied by alterations in the rate of mitochondrial Ca 2+ uptake 72. Conversely, MCUR1 knockdown in mammalian cells renders them resistant to Ca 2+ overload, providing protection from cell death 72. This led the authors to conclude that MCUR1 regulates the Ca 2+ threshold for the MPT and may act as a molecular bridge connecting the MCU channel to the MTP complex. Whether this hypothesis holds true still has to be tested.
More recently, the genetic deletion of MCUR1 in endothelial and cardiac tissues in vivo by mouse conditional knockout models 68 points to an involvement of MCUR1 in the assembly and function of the MCU channel. In particular, MCUR1 binding to MCU and EMRE appears to be necessary for MCU oligomerisation and stability, since MCUR1 –/– cells will display a significantly decreased amount of MCU-containing high-molecular-weight complexes and reduced rate of mitochondrial Ca 2+ uptake 68. This, as expected, impinges on cellular energetics, as it is reflected by the dramatic reduction of ATP levels and consequent activation of the pro-survival autophagy pathway in cells depleted of MCUR1 72. Whether the pleiotropic roles for MCUR1 in the regulation of mitochondrial Ca 2+ uptake, OXPHOS efficiency, and the apparent discordant phenotypes of MCUR1-depleted cells depend on the intrinsic transcriptional and metabolic differences of different cell types or on the species-specific features of the mitochondrial Ca 2+ machinery is still an open question, and further investigation is needed to clarify it.
The complexity of mitochondrial Ca 2+ uptake regulation
The regulation of the mitochondrial Ca 2+ uptake machinery has been revealed to be far more complex than that resulting from just considering the cooperative actions of the different MCU components and their possible combinations. Indeed, the activity of the MCU channel relies on a multifaceted integration of regulatory mechanisms acting on both the MCU pore subunits and its co-regulators. Moreover, these mechanisms have been shown to occur at multiple levels––transcriptional, post-transcriptional, and post-translational––and their systematic identification has started to be accomplished only in recent years.
In neurons, for example, MCU transcription appears to be under the control of activity-dependent cytosolic Ca 2+ signalling through the involvement of calmodulin (CaM) and the activation of CaM-kinase (CaMK) 73. It has been shown that the immediate-early gene Npas4, a neuronal transcription factor with neuroprotective action against excitotoxicity 74, acts directly downstream of the NMDAR signalling and of CaMK to modulate MCU transcription 73. In another context, chromatin immunoprecipitation and promoter reporter analyses revealed that the Ca 2+-regulated transcription factor cyclic adenosine monophosphate response element-binding protein (CREB) directly binds the MCU promoter and stimulates its transcriptional activity in chicken DT40 lymphocyte cells 75. The CREB-mediated activity-dependent modulation of MCU expression in response to intracellular Ca 2+ mobilisation via IP3R and store-operated Ca 2+ entry (SOCE) has also been proven to be crucial to ensure a prompt metabolic flexibility and cell survival in lymphocytes 75.
The existence of post-transcriptional regulation of MCU has been initially described in cancer 76, where a strong inverse correlation between MCU expression level and the abundance of microRNA (miR)-25 has been reported in both tumour cell lines and colon adenocarcinoma samples 76. Indeed, MCU expression is found to be upregulated in both colon and prostate cancer cells that show a reduced level of miR-25, and the overexpression of miR-25 in these cells downregulates MCU and increases cell sensitivity to apoptosis 76. Similarly, in breast cancer, the downregulation of miR-340 is correlated with increased MCU expression in highly metastatic cells, while MCU targeting by miR-340 blocks the metabolic shift from OXPHOS to aerobic glycolysis that would otherwise favour cell migration and invasiveness 77. In the context of pulmonary arterial hypertension (PAH), in which vascular cells are hyper-proliferative and apoptosis resistant, exhibiting a cancer-like phenotype, the decreased MCU function that underlies the key phenotypic features of PAH (including elevation of [Ca 2+] i, reduction of [Ca 2+] mt, mitochondrial fragmentation, and the Warburg phenomenon) has been ascribed to the downregulation of MCU expression by both miR-25 and miR-138 78. The overexpression of MCU or the blocking of the MCU-targeting miRs in PAH cells indeed restores Ca 2+ dynamics, mitochondrial metabolism, and cell proliferation to physiological levels 78. MiR-25 has also been shown to target and downregulate MCU in the heart, where it is suggested to protect myocytes from oxidative damage by reducing [Ca 2+] mt levels 79. Another two muscle-specific miRNAs (myomiR), miR-1 and miR-206, have been implicated in the regulation of MCU expression in muscle 80, of which miR-1 plays a major role in the context of cardiac development in both mice and humans. In mice, miR-1 upregulation is crucial during the first few postnatal weeks for the modulation of MCU activity in order to prevent massive [Ca 2+] mt elevation that would otherwise occur, since mitochondria distribution undergoes heavy remodelling at this time point and organelles are placed in close contact with the SR Ca 2+-releasing channels 80.
Post-translational modifications of MCU proteins have also been described. Indeed, the Ca 2+-dependent tyrosine kinase Pyk2 has been shown to directly interact and phosphorylate MCU in cardiac cells in response to α-adrenergic stimulation 81. The MCU phosphorylation by Pyk2 has been proposed to promote its oligomerisation and to enhance mitochondrial Ca 2+ entry, thus stimulating mitochondrial metabolism and favouring cytosolic Ca 2+ clearance in cardiac cells 81. In addition, the atomic resolution data of the N-terminus of MCU (residues 72–189) allowed the description of the exact structure of the MCU matrix domain 82. This domain was shown to acquire a β-grasp-like fold and to bind Mg 2+ and Ca 2+ through a MCU-regulating acidic patch (MRAP) 82. The disruption of cation binding via mutation of the MRAP sequence impairs MCU oligomerisation, reduces basal [Ca 2+] mt levels, and leads to significant dampening of mitochondrial Ca 2+ uptake after agonist stimulation, highlighting the role of divalent cations in the regulation of MCU activity 81. This has been proposed as a possible feedback mechanism of MCU regulation aimed at preventing excessive mitochondrial Ca 2+ entry in conditions of elevated [Ca 2+] i.
Finally, a wide range of chemical modifications have been reported to target the thiol moiety of Cys residues in many proteins, enabling biological switching of structure and reactivity oxidation 83. One of these modifications, S-glutathionylation, was recently shown to occur in MCU protein as the result of oxidation at the evolutionarily conserved Cys-97 84. The S-glutathionylation has been demonstrated to promote a MCU conformational change, which increases oligomerisation 84. In human pulmonary microvascular endothelial cells (HPMVECs), this enhances MCU activity and promotes increased mitochondrial Ca 2+ entry both in basal conditions and upon agonist challenge, causing cellular bioenergetics crisis and higher sensitivity to cell death 84.
Overall, the understanding of the mechanisms controlling Ca 2+ entry in mitochondria has made a great step forward in the last few years, and new possibilities for effective modulation of MCU activity have been revealed. In this line, very recently, a systematic orthologous interspecies chemical screen based on the combination of an optimised yeast cell system and a mammalian mitochondria-based NCC library drug screen picked out and validated the first MCU-specific inhibitor molecule to be described: mitoxantrone 85. Mitoxantrone has been utilised already in clinical practice for its antineoplastic action against non-Hodgkin’s lymphomas and acute myeloid leukaemia 86; however, the anti-tumour properties of this drug appear to rely on a different molecular moiety with respect to its anti-MCU activity, thus opening up the possibility for the chemical engineering of new lead compounds to specifically target MCU function. This is of outmost relevance for the design of novel potential therapeutic approaches to pathologies in which mitochondrial Ca 2+ signalling dysfunction is involved 17, 87, 88, including those characterised by primary MCU dysfunction due to mutations in its components 50, 52, 53.
In conclusion, despite many of the molecular mechanisms refining MCU activity in the cell having been described, of which the most relevant are presented in this review, a number of other possibilities for the modulation of uniporter function may exist (at transcriptional and post-transcriptional levels), and additional efforts are needed from the scientific community to fully unravel the complexity of mitochondrial Ca 2+ uptake regulation.
Conclusions
In the last few years, tremendous advances have been made in defining the molecular identity of the mitochondrial Ca 2+ transport machinery. Most of the components of the MCU complex have been identified, and several cellular and in vivo models have been generated that helped to define the physiological relevance of mitochondrial Ca 2+ uptake. Still, many issues remain obscure and need to be investigated. The entire spectrum of molecular mechanisms by which different cell types finely tune mitochondrial Ca 2+ uptake appears to be related not only to the metabolic needs but also to the breadth of cellular activities modulated by organelle Ca 2+ levels. We believe that the complete picture will emerge when at least three conceptual mechanisms are fully investigated, i.e. gene expression (the MCU and MICU1 isoforms show significant tissue-specific differences in expression), alternative splicing (as in the case of MICU1.1), and post-transcriptional and post-translational regulation (which, so far, has been only partially explored). We then need to explore the functional interplay between mitochondrial Ca 2+ transport and other ion fluxes, such as Na + and K +, for which complexity and partial redundancy of transport mechanisms have been described but molecular definition still lags behind. Unravelling this cross-talk will greatly enhance our insight into the regulation of mitochondria, these fascinating organelles with pleiotropic effects on a cell’s life and death.
Abbreviations
[Ca 2+] i, intracellular Ca 2+ concentration; [Ca 2+] mt, mitochondrial Ca 2+ concentration; CaM, calmodulin; CaMK, calmodulin kinase; CREB, cyclic adenosine monophosphate response element-binding protein; COX, cytochrome c oxidase; EMRE, essential mitochondrial Ca 2+ uniporter regulator; ER, endoplasmic reticulum; IMM, inner mitochondrial membrane; IMS, intermembrane space; IP3R, inositol trisphosphate receptor; MCU, mitochondrial Ca 2+ uniporter; MCUR1, mitochondrial Ca 2+ uniporter regulator 1; MICU, mitochondrial Ca 2+ uptake protein; miR, microRNA; MRAP, mitochondrial Ca 2+ uniporter-regulating acidic patch; MTP, mitochondrial permeability transition; OXPHOS, oxidative phosphorylation; PAH, pulmonary arterial hypertension; PGC1α4, peroxisome proliferator-activated receptor gamma coactivator 1 alpha 4; PTP, permeability transition pore; SR, sarcoplasmic reticulum; TMH, transmembrane helix
Author contributions
Giorgia Pallafacchina wrote the original draft, Sofia Zanin drew the figures, and Rosario Rizzuto revised the manuscript.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Dipayan Chaudhuri, Nora Eccles Harrison Cardiovascular Research and Training Institute, Cardiology Division, Department of Internal Medicine, University of Utah, Salt Lake City, UT, USA
Evgeny V Pavlov, Department of Basic Sciences, College of Dentistry, New York University, 345 East 24th Street, New York, NY, 10010, USA
Israel Sekler, Department of Physiology and Cell Biology, Faculty of Health Science, Ben-Gurion University, Beer-Sheva, Israel
Roberto Docampo, Center for Tropical and Emerging Global Diseases, University of Georgia, Athens, GA, 30602, USA
Funding Statement
This work was supported by grants to Rosario Rizzuto from the Italian Ministries of Health (Ricerca Finalizzata.) and of Education, University and Research (FIRB), the European Union (ERC mitoCalcium, no. 294777), the National Institutes of Health (grant #1P01AG025532-01A1), the Italian Association for Cancer Research (AIRC IG18633), and Telethon-Italy (GGP16029).
[version 1; referees: 4 approved]
References
- 1. Berridge MJ: Inositol trisphosphate and calcium signalling. Nature. 1993;361(6410):315–25. 10.1038/361315a0 [DOI] [PubMed] [Google Scholar]
- 2. Berridge MJ, Lipp P, Bootman MD: The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1(1):11–21. 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- 3. Clapham DE: Calcium signaling. Cell. 2007;131(6):1047–58. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- 4. Rizzuto R, De Stefani D, Raffaello A, et al. : Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13(9):566–78. 10.1038/nrm3412 [DOI] [PubMed] [Google Scholar]
- 5. Raffaello A, Mammucari C, Gherardi G, et al. : Calcium at the Center of Cell Signaling: Interplay between Endoplasmic Reticulum, Mitochondria, and Lysosomes. Trends Biochem Sci. 2016;41(12):1035–49. 10.1016/j.tibs.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berridge MJ: The versatility and complexity of calcium signalling. Novartis Found Symp. 2001;239:52–64; discussion 64–7, 150–9. [PubMed] [Google Scholar]
- 7. Calì T, Brini M, Carafoli E: Regulation of Cell Calcium and Role of Plasma Membrane Calcium ATPases. Int Rev Cell Mol Biol. 2017;332:259–96. 10.1016/bs.ircmb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 8. Vandecaetsbeek I, Vangheluwe P, Raeymaekers L, et al. : The Ca 2+ pumps of the endoplasmic reticulum and Golgi apparatus. Cold Spring Harb Perspect Biol. 2011;3(5): pii: a004184. 10.1101/cshperspect.a004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brini M, Calì T, Ottolini D, et al. : Calcium pumps: why so many? Compr Physiol. 2012;2(2):1045–60. 10.1002/cphy.c110034 [DOI] [PubMed] [Google Scholar]
- 10. Pinton P, Pozzan T, Rizzuto R: The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca 2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 1998;17(18):5298–308. 10.1093/emboj/17.18.5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calcraft PJ, Ruas M, Pan Z, et al. : NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459(7246):596–600. 10.1038/nature08030 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Deluca HF, Engstrom GW: Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci U S A. 1961;47:1744–50. 10.1073/pnas.47.11.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vasington FD, Murphy JV: Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–7. [PubMed] [Google Scholar]
- 14. Carafoli E, Rossi CS, Lehninger AL: Cation and Anion Balance During Active Accumulation of Ca++ and Mg++ by Isolated Mitochondria. J Biol Chem. 1964;239:3055–61. [PubMed] [Google Scholar]
- 15. Mitchell P: Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–8. 10.1038/191144a0 [DOI] [PubMed] [Google Scholar]
- 16. Penna E, Espino J, De Stefani D, et al. : The MCU complex in cell death. Cell Calcium. 2018;69:73–80. 10.1016/j.ceca.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 17. Granatiero V, De Stefani D, Rizzuto R: Mitochondrial Calcium Handling in Physiology and Disease. Adv Exp Med Biol. 2017;982:25–47. 10.1007/978-3-319-55330-6_2 [DOI] [PubMed] [Google Scholar]
- 18. Marchi S, Patergnani S, Missiroli S, et al. : Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium. 2018;69:62–72. 10.1016/j.ceca.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 19. Giacomello M, Drago I, Pizzo P, et al. : Mitochondrial Ca 2+ as a key regulator of cell life and death. Cell Death Differ. 2007;14(7):1267–74. 10.1038/sj.cdd.4402147 [DOI] [PubMed] [Google Scholar]
- 20. Kirichok Y, Krapivinsky G, Clapham DE: The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427(6972):360–4. 10.1038/nature02246 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Perocchi F, Gohil VM, Girgis HS, et al. : MICU1 encodes a mitochondrial EF hand protein required for Ca 2+ uptake. Nature. 2010;467(7313):291–6. 10.1038/nature09358 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Baughman JM, Perocchi F, Girgis HS, et al. : Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476(7360):341–5. 10.1038/nature10234 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. De Stefani D, Raffaello A, Teardo E, et al. : A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–40. 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Baradaran R, Wang C, Siliciano AF, et al. : Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018;559(7715):580–4. 10.1038/s41586-018-0331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Raffaello A, De Stefani D, Sabbadin D, et al. : The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32(17):2362–76. 10.1038/emboj.2013.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoo J, Wu M, Yin Y, et al. : Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018;361(6401):506–11. 10.1126/science.aar4056 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Nguyen NX, Armache JP, Lee C, et al. : Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature. 2018;559(7715):570–4. 10.1038/s41586-018-0333-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Fan C, Fan M, Orlando BJ, et al. : X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018;559(7715):575–9. 10.1038/s41586-018-0330-9 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Oxenoid K, Dong Y, Cao C, et al. : Architecture of the mitochondrial calcium uniporter. Nature. 2016;533(7602):269–73. 10.1038/nature17656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiurillo MA, Lander N, Bertolini MS, et al. : Different Roles of Mitochondrial Calcium Uniporter Complex Subunits in Growth and Infectivity of Trypanosoma cruzi. MBio. 2017;8(3): pii: e00574-17. 10.1128/mBio.00574-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang G, Docampo R: The Mitochondrial Ca 2+ Uniporter Complex (MCUC) of Trypanosoma brucei Is a Hetero-oligomer That Contains Novel Subunits Essential for Ca 2+ Uptake. MBio. 2018;9(5): pii: e01700-18. 10.1128/mBio.01700-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fieni F, Lee SB, Jan YN, et al. : Activity of the mitochondrial calcium uniporter varies greatly between tissues. Nat Commun. 2012;3:1317. 10.1038/ncomms2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barth E, Stämmler G, Speiser B, et al. : Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. J Mol Cell Cardiol. 1992;24(7):669–81. 10.1016/0022-2828(92)93381-S [DOI] [PubMed] [Google Scholar]
- 34. Pan X, Liu J, Nguyen T, et al. : The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat Cell Biol. 2013;15(12):1464–72. 10.1038/ncb2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murphy E, Pan X, Nguyen T, et al. : Unresolved questions from the analysis of mice lacking MCU expression. Biochem Biophys Res Commun. 2014;449(4):384–5. 10.1016/j.bbrc.2014.04.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwong JQ, Lu X, Correll RN, et al. : The Mitochondrial Calcium Uniporter Selectively Matches Metabolic Output to Acute Contractile Stress in the Heart. Cell Rep. 2015;12(1):15–22. 10.1016/j.celrep.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luongo TS, Lambert JP, Yuan A, et al. : The Mitochondrial Calcium Uniporter Matches Energetic Supply with Cardiac Workload during Stress and Modulates Permeability Transition. Cell Rep. 2015;12(1):23–34. 10.1016/j.celrep.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y, Rasmussen TP, Koval OM, et al. : The mitochondrial uniporter controls fight or flight heart rate increases. Nat Commun. 2015;6:6081. 10.1038/ncomms7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rasmussen TP, Wu Y, Joiner ML, et al. : Inhibition of MCU forces extramitochondrial adaptations governing physiological and pathological stress responses in heart. Proc Natl Acad Sci U S A. 2015;112(29):9129–34. 10.1073/pnas.1504705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nichols M, Elustondo PA, Warford J, et al. : Global ablation of the mitochondrial calcium uniporter increases glycolysis in cortical neurons subjected to energetic stressors. J Cereb Blood Flow Metab. 2017;37(8):3027–41. 10.1177/0271678X16682250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nichols M, Pavlov EV, Robertson GS: Tamoxifen-induced knockdown of the mitochondrial calcium uniporter in Thy1-expressing neurons protects mice from hypoxic/ischemic brain injury. Cell Death Dis. 2018;9(6):606. 10.1038/s41419-018-0607-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J, Wilhelmsson H, Graff C, et al. : Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21(1):133–7. 10.1038/5089 [DOI] [PubMed] [Google Scholar]
- 43. Sommakia S, Houlihan PR, Deane SS, et al. : Mitochondrial cardiomyopathies feature increased uptake and diminished efflux of mitochondrial calcium. J Mol Cell Cardiol. 2017;113:22–32. 10.1016/j.yjmcc.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mammucari C, Gherardi G, Zamparo I, et al. : The mitochondrial calcium uniporter controls skeletal muscle trophism in vivo. Cell Rep. 2015;10(8):1269–79. 10.1016/j.celrep.2015.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gherardi G, Nogara L, Ciciliot S, et al. : Loss of mitochondrial calcium uniporter rewires skeletal muscle metabolism and substrate preference. Cell Death Differ. 2018. 10.1038/s41418-018-0191-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mallilankaraman K, Doonan P, Cárdenas C, et al. : MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca 2+ uptake that regulates cell survival. Cell. 2012;151(3):630–44. 10.1016/j.cell.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Csordás G, Golenár T, Seifert EL, et al. : MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca 2+ uniporter. Cell Metab. 2013;17(6):976–87. 10.1016/j.cmet.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu JC, Liu J, Holmström KM, et al. : MICU1 Serves as a Molecular Gatekeeper to Prevent In Vivo Mitochondrial Calcium Overload. Cell Rep. 2016;16(6):1561–73. 10.1016/j.celrep.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Antony AN, Paillard M, Moffat C, et al. : MICU1 regulation of mitochondrial Ca 2+ uptake dictates survival and tissue regeneration. Nat Commun. 2016;7:10955. 10.1038/ncomms10955 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Logan CV, Szabadkai G, Sharpe JA, et al. : Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat Genet. 2014;46(2):188–93. 10.1038/ng.2851 [DOI] [PubMed] [Google Scholar]
- 51. Kamer KJ, Sancak Y, Fomina Y, et al. : MICU1 imparts the mitochondrial uniporter with the ability to discriminate between Ca 2+ and Mn 2+ Proc Natl Acad Sci U S A. 2018;115(34):E7960–E7969. 10.1073/pnas.1807811115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Lewis-Smith D, Kamer KJ, Griffin H, et al. : Homozygous deletion in MICU1 presenting with fatigue and lethargy in childhood. Neurol Genet. 2016;2(2):e59. 10.1212/NXG.0000000000000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Musa S, Eyaid W, Kamer K, et al. : A Middle Eastern Founder Mutation Expands the Genotypic and Phenotypic Spectrum of Mitochondrial MICU1 Deficiency: A Report of 13 Patients. JIMD Rep. 2018;1–5. 10.1007/8904_2018_107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Plovanich M, Bogorad RL, Sancak Y, et al. : MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One. 2013;8(2):e55785. 10.1371/journal.pone.0055785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Patron M, Granatiero V, Espino J, et al. : MICU3 is a tissue-specific enhancer of mitochondrial calcium uptake. Cell Death Differ. 2018. 10.1038/s41418-018-0113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Patron M, Checchetto V, Raffaello A, et al. : MICU1 and MICU2 finely tune the mitochondrial Ca 2+ uniporter by exerting opposite effects on MCU activity. Mol Cell. 2014;53(5):726–37. 10.1016/j.molcel.2014.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kamer KJ, Grabarek Z, Mootha VK: High-affinity cooperative Ca 2+ binding by MICU1-MICU2 serves as an on-off switch for the uniporter.. EMBO Rep. 2017;18(8):1397–411. 10.15252/embr.201643748 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Payne R, Hoff H, Roskowski A, et al. : MICU2 Restricts Spatial Crosstalk between InsP 3R and MCU Channels by Regulating Threshold and Gain of MICU1-Mediated Inhibition and Activation of MCU. Cell Rep. 2017;21(11):3141–54. 10.1016/j.celrep.2017.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Paillard M, Csordás G, Szanda G, et al. : Tissue-Specific Mitochondrial Decoding of Cytoplasmic Ca 2+ Signals Is Controlled by the Stoichiometry of MICU1/2 and MCU. Cell Rep. 2017;18(10):2291–300. 10.1016/j.celrep.2017.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Vecellio Reane D, Vallese F, Checchetto V, et al. : A MICU1 Splice Variant Confers High Sensitivity to the Mitochondrial Ca 2+ Uptake Machinery of Skeletal Muscle. Mol Cell. 2016;64(4):760–73. 10.1016/j.molcel.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 61. Sancak Y, Markhard AL, Kitami T, et al. : EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342(6164):1379–82. 10.1126/science.1242993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vais H, Mallilankaraman K, Mak DD, et al. : EMRE Is a Matrix Ca 2+ Sensor that Governs Gatekeeping of the Mitochondrial Ca 2+ Uniporter. Cell Rep. 2016;14(3):403–10. 10.1016/j.celrep.2015.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Tsai MF, Phillips CB, Ranaghan M, et al. : Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife. 2016;5: pii: e15545. 10.7554/eLife.15545 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Yamamoto T, Yamagoshi R, Harada K, et al. : Analysis of the structure and function of EMRE in a yeast expression system. Biochim Biophys Acta. 2016;1857(6):831–9. 10.1016/j.bbabio.2016.03.019 [DOI] [PubMed] [Google Scholar]
- 65. König T, Tröder SE, Bakka K, et al. : The m-AAA Protease Associated with Neurodegeneration Limits MCU Activity in Mitochondria. Mol Cell. 2016;64(1):148–62. 10.1016/j.molcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 66. Patron M, Sprenger HG, Langer T: m-AAA proteases, mitochondrial calcium homeostasis and neurodegeneration. Cell Res. 2018;28(3):296–306. 10.1038/cr.2018.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mallilankaraman K, Cárdenas C, Doonan PJ, et al. : MCUR1 is an essential component of mitochondrial Ca 2+ uptake that regulates cellular metabolism. Nat Cell Biol. 2012;14(12):1336–43. 10.1038/ncb2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tomar D, Dong Z, Shanmughapriya S, et al. : MCUR1 Is a Scaffold Factor for the MCU Complex Function and Promotes Mitochondrial Bioenergetics. Cell Rep. 2016;15(8):1673–85. 10.1016/j.celrep.2016.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Paupe V, Prudent J, Dassa EP, et al. : CCDC90A (MCUR1) is a cytochrome c oxidase assembly factor and not a regulator of the mitochondrial calcium uniporter. Cell Metab. 2015;21(1):109–16. 10.1016/j.cmet.2014.12.004 [DOI] [PubMed] [Google Scholar]
- 70. Sickmann A, Reinders J, Wagner Y, et al. : The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100(23):13207–12. 10.1073/pnas.2135385100 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Vais H, Tanis JE, Müller M, et al. : MCUR1, CCDC90A, Is a Regulator of the Mitochondrial Calcium Uniporter. Cell Metab. 2015;22(4):533–5. 10.1016/j.cmet.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chaudhuri D, Artiga DJ, Abiria SA, et al. : Mitochondrial calcium uniporter regulator 1 (MCUR1) regulates the calcium threshold for the mitochondrial permeability transition. Proc Natl Acad Sci U S A. 2016;113(13):E1872–80. 10.1073/pnas.1602264113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qiu J, Tan YW, Hagenston AM, et al. : Mitochondrial calcium uniporter Mcu controls excitotoxicity and is transcriptionally repressed by neuroprotective nuclear calcium signals. Nat Commun. 2013;4:2034. 10.1038/ncomms3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang SJ, Zou M, Lu L, et al. : Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5(8):e1000604. 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shanmughapriya S, Rajan S, Hoffman NE, et al. : Ca 2+ signals regulate mitochondrial metabolism by stimulating CREB-mediated expression of the mitochondrial Ca 2+ uniporter gene MCU. Sci Signal. 2015;8(366):ra23. 10.1126/scisignal.2005673 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Marchi S, Lupini L, Patergnani S, et al. : Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25. Curr Biol. 2013;23(1):58–63. 10.1016/j.cub.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yu C, Wang Y, Peng J, et al. : Mitochondrial calcium uniporter as a target of microRNA-340 and promoter of metastasis via enhancing the Warburg effect. Oncotarget. 2017;8(48):83831–44. 10.18632/oncotarget.19747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hong Z, Chen KH, DasGupta A, et al. : MicroRNA-138 and MicroRNA-25 Down-regulate Mitochondrial Calcium Uniporter, Causing the Pulmonary Arterial Hypertension Cancer Phenotype. Am J Respir Crit Care Med. 2017;195(4):515–29. 10.1164/rccm.201604-0814OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pan L, Huang BJ, Ma XE, et al. : MiR-25 protects cardiomyocytes against oxidative damage by targeting the mitochondrial calcium uniporter. Int J Mol Sci. 2015;16(3):5420–33. 10.3390/ijms16035420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zaglia T, Ceriotti P, Campo A, et al. : Content of mitochondrial calcium uniporter (MCU) in cardiomyocytes is regulated by microRNA-1 in physiologic and pathologic hypertrophy. Proc Natl Acad Sci U S A. 2017;114(43):E9006–E9015. 10.1073/pnas.1708772114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. O-Uchi J, Jhun BS, Xu S, et al. : Adrenergic signaling regulates mitochondrial Ca 2+ uptake through Pyk2-dependent tyrosine phosphorylation of the mitochondrial Ca 2+ uniporter. Antioxid Redox Signal. 2014;21(6):863–79. 10.1089/ars.2013.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee SK, Shanmughapriya S, Mok MCY, et al. : Structural Insights into Mitochondrial Calcium Uniporter Regulation by Divalent Cations. Cell Chem Biol. 2016;23(9):1157–69. 10.1016/j.chembiol.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Wani R, Nagata A, Murray BW: Protein redox chemistry: Post-translational cysteine modifications that regulate signal transduction and drug pharmacology. Front Pharmacol. 2014;5:224. 10.3389/fphar.2014.00224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dong Z, Shanmughapriya S, Tomar D, et al. : Mitochondrial Ca 2+ Uniporter Is a Mitochondrial Luminal Redox Sensor that Augments MCU Channel Activity. Mol Cell. 2017;65(6):1014–1028.e7. 10.1016/j.molcel.2017.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Arduino DM, Wettmarshausen J, Vais H, et al. : Systematic Identification of MCU Modulators by Orthogonal Interspecies Chemical Screening. Mol Cell. 2017;67(4):711–723.e7. 10.1016/j.molcel.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Evison BJ, Sleebs BE, Watson KG, et al. : Mitoxantrone, More than Just Another Topoisomerase II Poison. Med Res Rev. 2016;36(2):248–99. 10.1002/med.21364 [DOI] [PubMed] [Google Scholar]
- 87. Mammucari C, Gherardi G, Rizzuto R: Structure, Activity Regulation, and Role of the Mitochondrial Calcium Uniporter in Health and Disease. Front Oncol. 2017;7:139. 10.3389/fonc.2017.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Calì T, Ottolini D, Brini M: Mitochondrial Ca 2+ and neurodegeneration. Cell Calcium. 2012;52(1):73–85. 10.1016/j.ceca.2012.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]