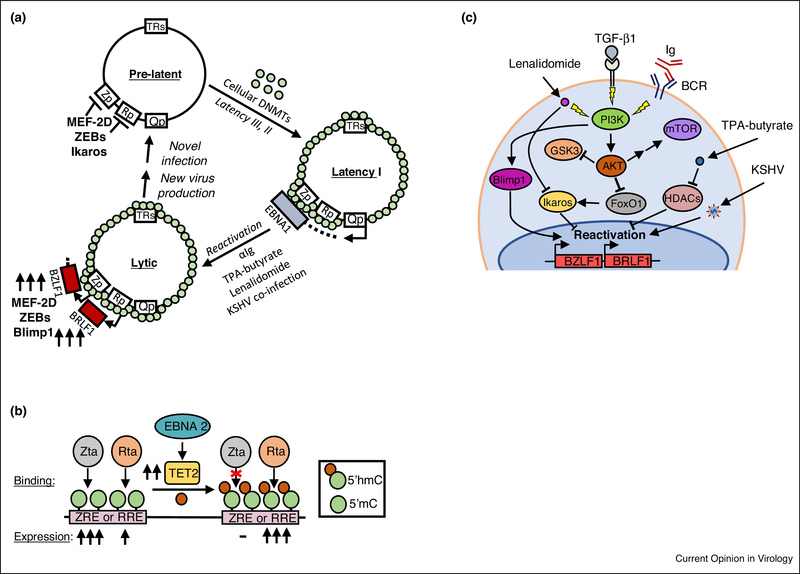
Figure 2. Epigenetic mechanisms affecting the EBV latent-lytic switch.
(A) Schematic diagram depicting repressive methylation (green circles) accrual on the EBV genome by cellular DNA methyltransferases (DMNTs), resulting in latency I. Shown also are selected treatments that can induce lytic gene expression. (B) TET2 conversion of viral genome methylated Z-response element (ZRE) 5’cytosine residues (5mC) into 5’ hydroxymethylation (5’hmC) precludes Zta binding and transactivation. 5’hmC does not affect Rta binding, but surprisingly enhances Rta-mediated activation. (C) PI3K pathway activation stimulates EBV lytic reactivation from latency I. Multiple lytic reactivation stimuli activate PI3K to upregulate Blimp1 and downmodulate Ikaros.