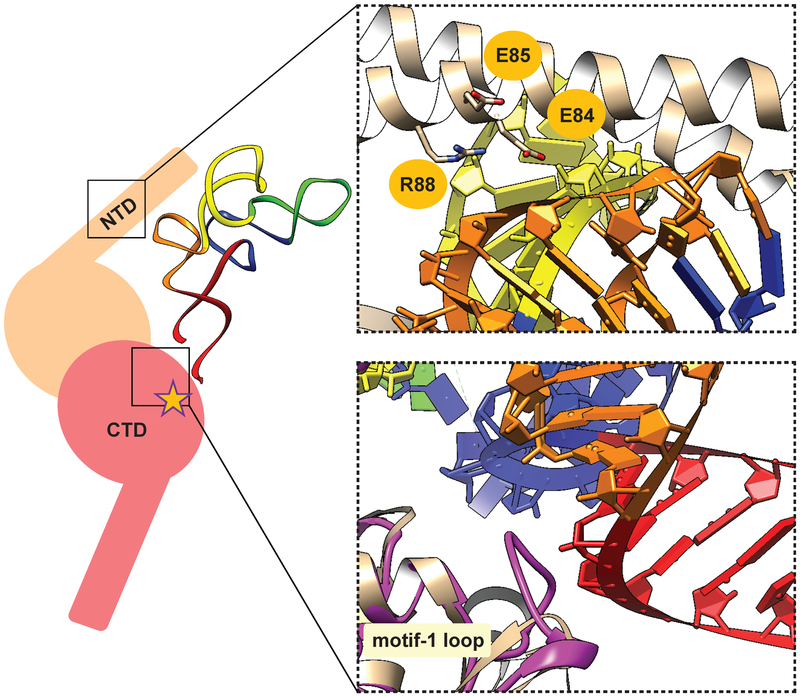
Fig. 2.
Key interactions between SerRS and tRNASer as a guide for SerRS engineering. (left) Cartoon representation of SerRS•tRNASec productive complex. SerRS is given in two shades of pink, for each monomer of the homodimer. The N-terminal tRNA binding domain (NTD), C-terminal catalytic domain (CTD) and the active site (star) are indicated. Cognate tRNA binds across the dimer. Acceptor, TψC, variable, anticodon, and D arms of tRNASec are shown in red, orange, yellow, green and blue, respectively. (right) Magnified view of the two regions targeted for mutagenesis. (up) In the crystal structure of T. thermophilus SerRS•tRNASer complex (PDB ID: 1SER) NTD residues E84, E85, and R88 are in the vicinity and/or form contacts with the TψC arm. These residues correspond to K86, A87, and D90 of E. coli SerRS. (down) Compared to T. thermophilus SerRS (light pink), E. coli SerRS (purple) has a longer motif 1 loop, which may interact with the base of the acceptor arm.