Fig 5. Summary of mutation types identified by multi-level structural analysis.
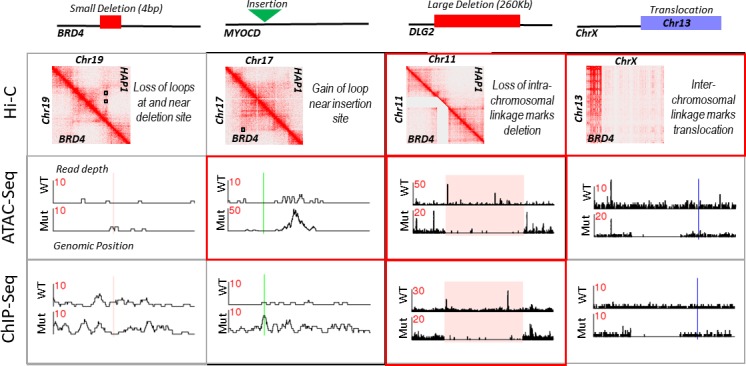
Impact of the four different mutation types that were agnostically identified through our holistic analysis on secondary and tertiary DNA structures. All four mutations are found in the BRD4 mutant. The name listed under the mutation type is the gene or chromosome impacted by this mutation. ATAC- and ChIP-Seq read depth traces are as described in Fig 3. Hi-C contact maps for the left three mutation types show data for the modified strain in the lower left portion of the heat map, and for the un-modified strain in the upper right portion. Hi-C contact data for the translocation is only shown for the mutant strain. Black boxes designate loops called for each respective data sets. Colored boxes on the read traces designate the deleted region (red), insertion site (green), or putative chromosomal breakpoint (blue). Boxes that have a red border designate platforms where there structural change was significantly different as determined by differential analysis. ChIP-Seq data is for the H3K27Ac antibody only. Expanded analyses for these mutations are given in Fig 3, S5 Fig, S3 Fig, and S6 Fig.
