Figure 1.
Identification of New lncRNAs during Embryonic Development
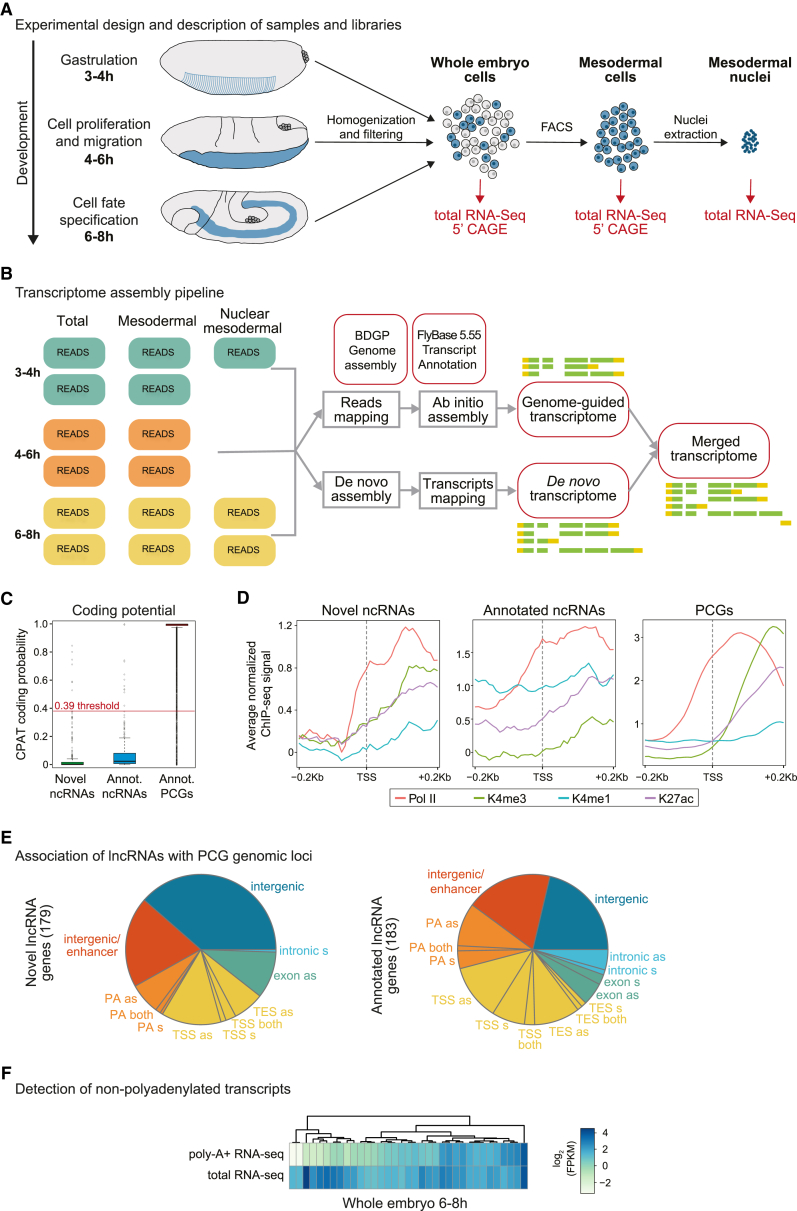
(A) Schematic overview of experimental design. Whole-embryo and mesodermal total RNA-seq stranded (100 bp paired-end) and 5′ CAGE (50 bp single-end) libraries were sequenced from 3–4 hr, 4–6 hr, and 6–8 hr embryos. Mesodermal nuclear RNA-seq libraries were prepared from 3–4 hr and 6–8 hr.
(B) Strategy overview of transcriptome assembly, combining ab initio and de novo assembly (STAR Methods).
(C) Novel lncRNAs lack coding potential. Boxplot showing CPAT coding potential predictions for our novel lncRNAs, previously annotated lncRNAs (FlyBase 5.55 annotation) and protein-coding genes (PCGs). Red line indicates threshold for coding potential (0.39).
(D) Histone modifications and RNA polymerase II (Pol II) presence at transcript start sites. Average chromatin immunoprecipitation sequencing (ChIP-seq) signal is shown for H3K27ac, H3K4me1, H3K4me3, and Pol II in mesoderm from 6–8 hr embryos [43], for promoter regions of novel and annotated lncRNAs and PCGs.
(E) Pie charts showing the genomic distribution of novel and annotated lncRNA genes with respect to PCGs. Genes are assigned to one class following the hierarchy: TSS > TES > exon > intron > promoter > enhancer > intergenic.
(F) Polyadenylation status of lncRNAs. Heatmaps show expression levels of novel lncRNAs in total RNA-seq (ribodepleted) and polyA-selected RNA-seq libraries from matched 6–8 hr whole-embryo samples.