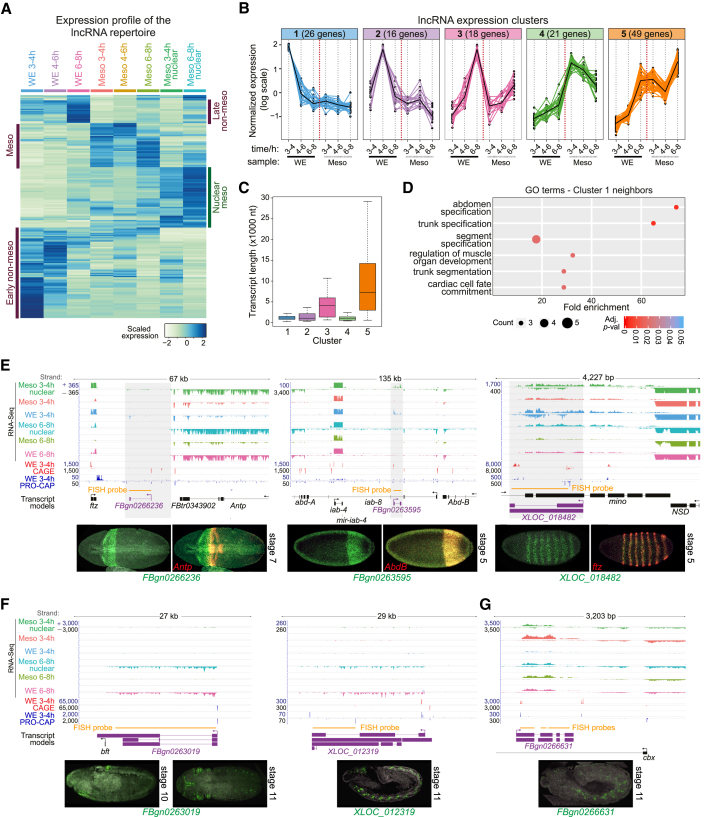
Figure 2.
Expression Properties of Developmental lncRNAs
(A) Heatmap showing scaled expression levels of 362 lncRNA genes (both novel and annotated) across 8 conditions. Groups with mesodermal and nuclear over-or under-expression are indicated.
(B) Iterative k-means clustering (STAR Methods) identifies five robust groups of lncRNAs with highly correlated expression across conditions. x axis denotes the experimental conditions and y axis the normalized, scaled gene expression levels.
(C) Boxplot showing size distribution of transcripts in each expression cluster. Early clusters (1, 2, and 4) are significantly smaller than late clusters (p = 1.294 × 10−12; Wilcoxon test).
(D) Protein coding genes in the vicinity of early cluster 1 have functions in early embryo patterning. Dot plot shows GO biological process term enrichment for the two closest PCGs (one neighbor either side of each cluster 1 gene). x axis indicates fold enrichment between observed and expected and y axis the significant terms sorted by decreasing p value. Dot size reflects the number of genes in that ontology, and dot color indicates p value, corrected for multiple testing. Uncorrected p values for all significant terms in all clusters are shown in Figure S5.
(E–G) (Above) Genomic regions showing lncRNA expression (purple gene models) and their close neighbors (black gene models) across samples. The direction of transcription is indicated by reads above (sense) or below (antisense) the lines. Meso, mesoderm from FACS-purified cells; WE, whole embryo. (Below) Fluorescence in situ hybridization (FISH) images of lncRNA show early expression patterns (E), late expression patterns (F), or belonging to mesoderm-enriched set (G).
See also Figures S4, S5, and S6 and Table S5.