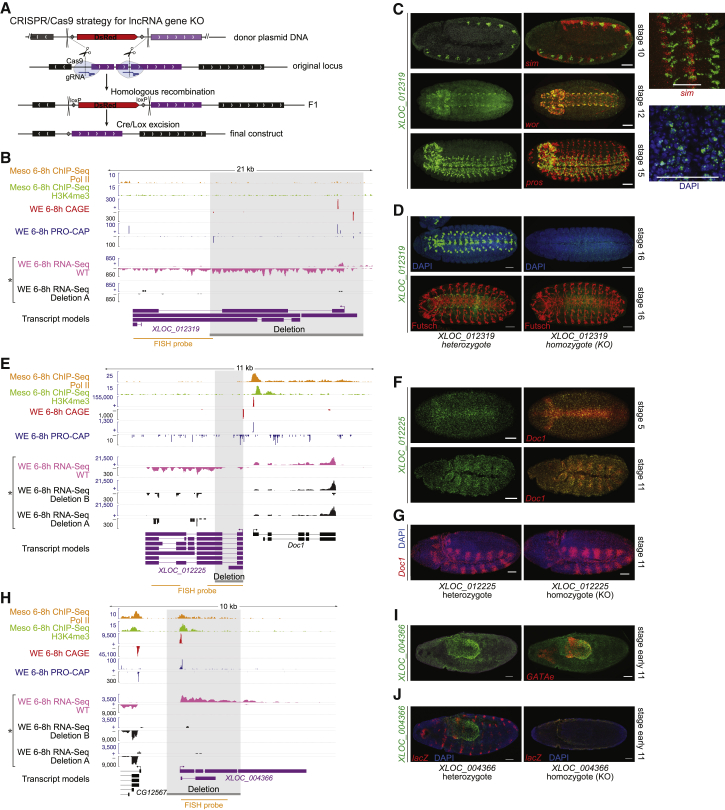
Figure 4.
Assessing lncRNA Function by Gene Knockout
(A) Strategy for genetic deletions with CRISPR/Cas9 [54].
(B, E, and H) Genomic loci of XLOC_012319 (B), XLOC_012225 (E), and XLOC_004366 (H) (purple; gene model) showing RNA-seq in WEs (pink track) and homozygous mutants from the deleted line (black track) at 6–8 hr, indicated by asterisk. Deleted region indicated by gray shading.
(C) Spatiotemporal expression of XLOC_012319. (Left panels) Double FISH of lncRNA (green; upper embryos) with different neuronal marker genes (red): ventral midline marker single-minded (sim); neuroblast marker worniu (wor); and ganglion mother cell (GMC) marker prospero (pros). (Right panels, upper) Zoomed image of XLOC_012319 and sim expression shows co-expression in ventral midline. Signals do not overlap perfectly as XLOC_012319 RNA is predominantly nuclear, shown by co-staining with DAPI.
(D) FISH of heterozygous and homozygous XLOC_012319 embryos. XLOC_012319 deletion completely abolishes lncRNA expression (left) but does not obviously affect neuronal development, as seen by immunofluorescence with an antibody against the neuronal marker Futsch (right).
(F) Double FISH of XLOC_012225 lncRNA (green) and its divergent PCG, Dorsocross-1 (Doc1, red). Left, stage 5 embryo, dorsal view, and right, stage 11 lateral view, show highly overlapping expression in dorsal ectoderm and amnioserosa.
(G) XLOC_012225 KO (homozygous embryos) has normal Doc1 expression.
(I) XLOC_004366 is detected at low levels throughout the embryo and enriched in posterior endoderm primordium (green), marked by GATAe expression (red).
(J) lncRNA expression is undetectable in the homozygous mutant embryos (right). Heterozygous embryos were identified by lacZ expression from the balancer chromosome (red).
All scale bars represent 50 μm. See also Table 1.