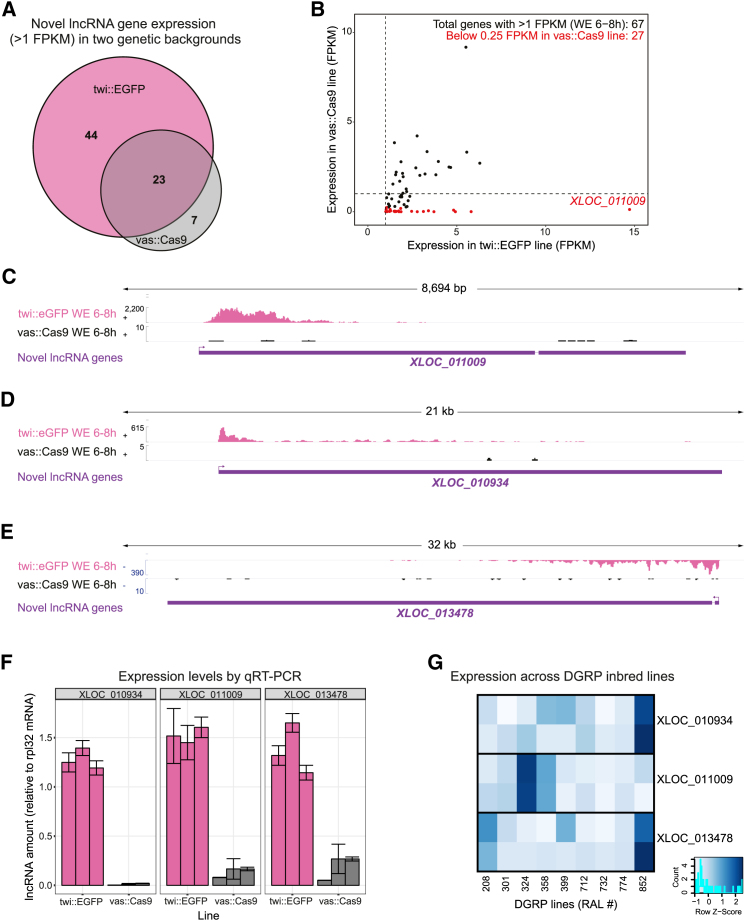
Figure 6.
Detection of Strain-Specific Transcripts
(A) Venn diagram showing intersection between transcripts expressed (>1 FPKM) in two genetic backgrounds (twi::EGFP and vas::Cas9). Unique transcripts are biased toward the twi::EGFP line as the transcriptome assembly is based on this line.
(B) Expression values for the 67 lncRNA genes with expression >1 FPKM in the twi::EGFP background. Scatterplot indicates that 27 genes are virtually not expressed in vas::Cas9 line (red dots).
(C–E) XLOC_011009 (C), XLOC_010934 (D), and XLOC_013478 (E) are examples of lncRNAs expressed in the twi::EGFP genetic background but completely absent at matching embryonic stages from the vas::Cas9 background.
(F) qRT-PCR confirming differences between twi::EGFP and vas::Cas9. Each bar represents an independent biological replicate; error bar is SE (from reaction duplicates).
(G) Assessment of strain-specific lncRNA expression across nine inbred Drosophila lines. Heatmap indicates qRT-PCR values. For each transcript line combination, the two tiles correspond to biological replicates.