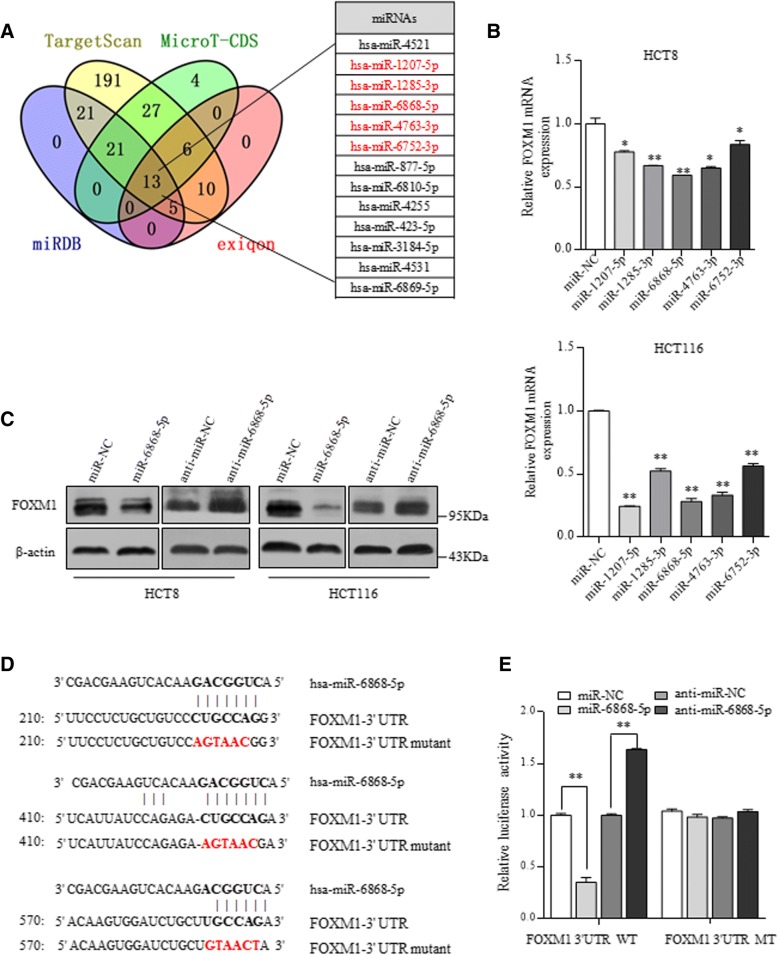
Fig. 1.
FOXM1 is negatively regulated by miR-6868-5p. a FOXM1 3’-UTR region was used for analysis of miRNA binding sites by using four different bioinformatic tools. miRNAs with at least 3 putative binding sites in at least two prediction tools were marked in red. b qRT-PCR analysis of FOXM1 expression in HCT8 and HCT116 cells transfected with indicated miRNA mimics. c Western blot analysis of FOXM1 expression in HCT8 and HCT116 cells transfected with miR-6868-5p mimic or miR-6868-5p inhibitor. d Locations of miR-6868-5p binding sites in the FOXM1 3’-UTR and the mutated FOXM1 3’-UTR are shown. e Luciferase activity of the wild-type (WT) or mutant (MT) FOXM1 3’-UTR reporter was measured in HCT116 cells transfected with miR-6868-5p mimic or inhibitor. *p < 0.05, **p < 0.01