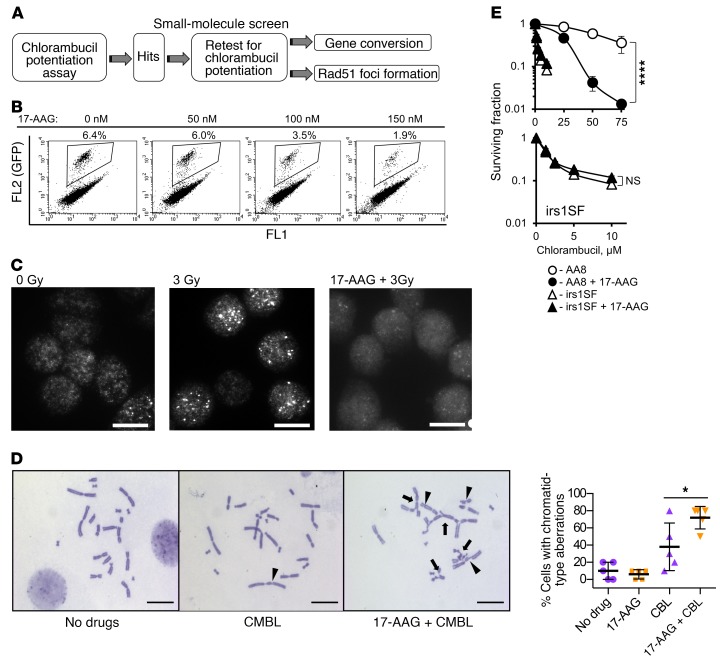
Figure 1. Overview of the small-molecule screen performed to identify inhibitors of homology-directed repair (HDR).
(A) Diagram of the screen. (B–E) 17-Allylamino-17-demethoxygeldanamycin (17-AAG) is used as a positive control for the screen. (B) 17-AAG inhibits gene conversion in the U2OS-DR-GFP cells. Details on gene conversion assay and quantification are provided in Supplemental Figure 1, A and C. (C) 17-AAG (100 nM) inhibits formation of Rad51 foci in the CHO AA8 cells after 3 Gy. Images were taken at 2 hours after irradiation. Representative images from n ≥ 3 experiments are shown. Scale bars: 10 μm. Quantification of signals is provided in Figure 2D. (D) Chlorambucil (CMBL; 5 μM) induces chromatid-type aberrations in CHO AA8 cells, and 17-AAG (150 nM) potentiates this effect. Arrowheads point to chromatid gaps and breaks, and arrows to complex chromatid exchanges. Scale bars: 20 μm. Graph on the right shows quantitation for data exemplified on the left. Significance analysis: 2-way ANOVA (P = 0.0343). Distribution of chromatid-type aberrations for each treatment is shown in Supplemental Figure 1E. (E) 17-AAG (50 nM) increases sensitivity of CHO AA8 cells to chlorambucil, but does not affect sensitivity of HDR-deficient CHO irs1SF cells, as measured by MTS assay. Bottom: The same data as in the top panel for the irs1SF cells at lower concentrations of chlorambucil. Shown are means ± SDs from n ≥ 3 experiments. Significance analysis: 2-way ANOVA (P < 0.0001). *P < 0.05, ****P < 0.0001.