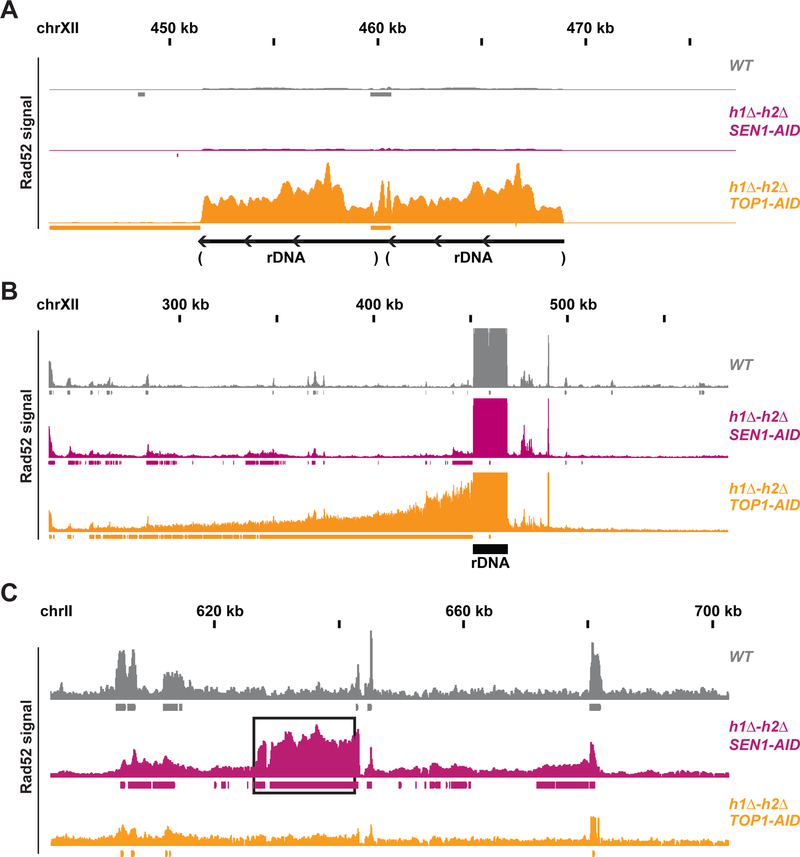
Fig. 4. Rad52-ChIPseq identifies rDNA as region of DNA damage in h1Δ-h2Δ TOP1-AID.
(A) Rad52-ChIPseq signal from WT(grey), h1Δ-h2Δ SEN1-AID (purple) and h1Δ-h2Δ TOP1-AID (orange) on the ribosomal-DNA locus (rDNA), after auxin treatment to deplete Sen1-AID and Top1-AID. Just two repeats of the rDNA are present in the annotated genome, while cells possess around 150 copies of the rDNA. Upon Top1 depletion, h1Δ-h2Δ TOP1-AID cells present a 20-fold increase in Rad52 signal on the rDNA region compared to other strains. (B) Rad52-ChIPseq signal from a larger portion of chromosome XII containing the rDNA locus. Rad52 signal from strains as in (A). The signal on the repetitive rDNA locus is around 75-fold higher than the one from unique regions; because the reference genome used in the alignment has just 2 rDNA copies while cells possess around 150 copies. By lowering the maximum signal by around 75 folds and showing a larger portion of the chromosome XII with the unique sequence flanking the rDNA, we detected a new Rad52 region large 150kb on the centromere-side of the rDNA, but not in the telomere side. H1Δ-h2Δ TOP1-AID presents a new Rad52 region in the rDNA and in the 150kb on the centromere-flanking unique region. (C) Rad52 signal from strains as in (A). A representative new Rad52 region identified in h1Δ-h2Δ SEN1-AID (black box) is not present in h1Δ-h2Δ TOP1-AID cells.