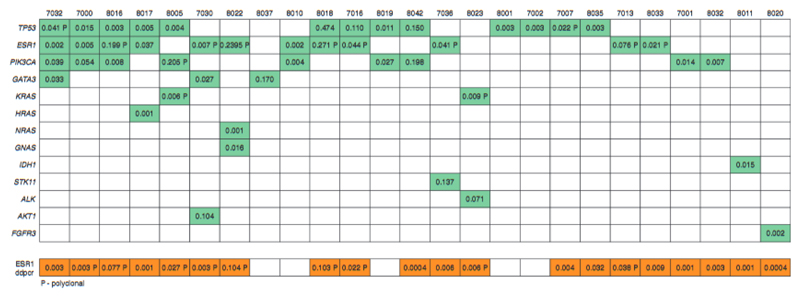
Figure 3.
Error corrected ctDNA sequencing of plasma samples taken after progression on the first-line aromatase inhibitor (AI). Mutations identified in plasma DNA by eTAm-Seq error corrected sequencing, with ESR1 mutation analysis by ddPCR. Discordant cases for ESR1 between ddPCR ard ctDNA sequencing had lower mutant copies per ml in ddPCR compared with concordant cases (median 14.3 versus 51.5, respectively, P = 0.048 Mann–Whitney U test) and likely represent very low levels of mutant copies and random sampling. 8037 also had FGFR1 and ERBB2 amplification Identified. Of 36 progression plasma samples sequenced, 25 with mutations are displayed, 11 plasma samples with no mutations detected are not displayed. Numbers in boxes represent allele fraction for indicated gene. Where there are multiple mutations detected in the same gene, indicating polyclonality (P), aggregate allele fractions are given.