Abstract
N-terminal acetylation (NTA) is a post-transcriptional modification of proteins that is conserved from bacteria to humans. In bacteria, the enzymes that mediate protein NTA also promote antimicrobial resistance. In pathogenic mycobacteria, which cause human tuberculosis and other chronic infections, NTA has been linked to pathogenesis and stress response, yet the fundamental biology underlying NTA of mycobacterial proteins remains unclear. We enriched, defined, and quantified the NT-acetylated populations of both cell-associated and secreted proteins from both the human pathogen, Mycobacterium tuberculosis, and the nontuberculous opportunistic pathogen, Mycobacterium marinum. We used a parallel N-terminal enrichment strategy from proteolytic digests coupled to charge-based selection and stable isotope ratio mass spectrometry. We show that NTA of the mycobacterial proteome is abundant, diverse, and primarily on Thr residues, which is unique compared with other bacteria. We isolated both the acetylated and unacetylated forms of 256 proteins, indicating that NTA of mycobacterial proteins is homeostatic. We identified 16 mycobacterial proteins with differential levels of NTA on the cytoplasmic and secreted forms, linking protein modification and localization. Our findings reveal novel biology underlying the NTA of mycobacterial proteins, which may provide a basis to understand NTA in mycobacterial physiology, pathogenesis, and antimicrobial resistance.
Keywords: N-terminal acetylation, tuberculosis, N-terminomics, N-terminal enrichment, bacterial proteomics, mycobacteria
Graphical Abstract:
INTRODUCTION
N-terminal acetylation (NTA) is addition of an acyl group to the free α-amino group of the terminal amino acid of a polypeptide. This is predominately catalyzed by N-acetyl transferases (NATs) with acetyl Co-A as the acetyl donor.1 NTA is distinct from ε-amino acetylation of lysine residues.1 NTA of proteins in higher organisms is abundant. In yeast and humans, NTA occurs on 70 and 90% of the observable N-terminal proteome, respectively.2–5 Although the precise role of NTA remains unknown, NTA has been linked to mediating protein interactions, turnover, stability, and localization.6–9 NTA has been reported at a lower extent in bacterial proteomes. Studies of NTA in bacteria initially suggested that NTA was limited to ribosomal proteins.10–14 Recently, more comprehensive studies in bacteria have vastly extended the number and categories of proteins with NTA. Importantly, NTA occurs on ~10% of detected proteins, or ~3% of the entire predicted proteome, in the three pathogens, Acinetobacter baummanii, Pseudomonas aeruginosa, and Escherichia coli.15–19
Mycobacterium tuberculosis (M. tb) is the causative agent of the human disease tuberculosis (TB).20 TB is the leading cause of death by an infectious disease, killing more than 1.7 million people globally in 2016.21,22 In addition to mycobacterial species that cause TB, there are several nontuberculous mycobacteria (NTM) that are an emerging health threat.23,24 Mycobacterium marinum is an NTM and an opportunistic human pathogen and an established model for studying M. tb pathogenesis.25–29 A large-scale, shotgun proteogenomic approach aimed at improving genomic annotation reported NTA of proteins in the H37Rv laboratory strain of M. tb.30 However, a focused analysis and method for NT-acetylated proteins was not performed.
Whereas the major function of NTA of bacterial proteomes remains unclear, NTA and the NATs that mediate this modification have been linked to several critical processes in mycobacteria. In addition to proteins, NATs acetylate antimicrobial drugs, including aminoglycosides, which often leads to antimicrobial resistance.31,32 For example, the AAC(6′)-ly NAT, which is conserved in Salmonella and Mycobacterium, is capable of both protein N-α acetylation and acetylation of antibiotics.31 Moreover, NAT activity is essential for the formation of mycothiol, a glutathione-like molecule produced by mycobacteria that maintains redox state in the cytosol and detoxifies cells during oxidative stress.32,33 We and others have reported NTA of virulence factors secreted by ESX systems,30,34–41 which promote the survival of the bacteria within host macrophages.42–46
Despite the reports of NTA in mycobacteria, the extent to which the homeostasis of acetylated proteins influences antibiotic acetylation, detoxification, and pathogenesis is unknown. As such, a quantitative census of NTA in mycobacteria is essential to understanding its role in cellular physiology and pathogenesis. Moreover, understanding NTA of mycobacterial proteins will enable characterization of drug resistance mechanisms.
There are several challenges to understanding the fundamental biology underlying NTA. The occurrence of NTA cannot be predicted based on the amino acid sequence of a protein. Therefore, the measurement of protein N-termini is required to define the scope of NTA in proteomes.47–51 Direct measurement of protein N-termini through “N-terminomic strategies” has been applied to bottom-up and top-down proteomics approaches.49–52 In bottom-up approaches, the presence of internal neo-peptides, which result from cleavage by proteases including trypsin, cause N-termini to be relatively underrepresented. The low representation of N-termini in bottom-up proteomes makes identification and analysis of N-termini challenging. Top-down strategies are not routine and are less sensitive than peptide-based approaches.53
N-terminal enrichment strategies increase the relative proportion of the natural N-termini in a proteome. These methods for intact or digested proteins can be generally categorized into either positive or negative enrichment. Positive enrichment methods typically biochemically enrich N-termini by selective affinity or capture of N-terminal peptides. Negative enrichment techniques deplete unmodified N-termini and neo-peptides.48,54 COmbined FRactional DIagonal Chromatography (COFRADIC), a negative enrichment approach, has distinguished N-termini in higher organisms by using orthogonal reverse-phase chromatography, followed by LC−MS/MS analysis of enriched peptide fractions.55 In general, enrichment is achieved by chemically blocking protein N-termini, digesting the proteomes into peptides, and depleting the neo-peptides, which contain a new (neo) free primary amine.55–60 Identification is performed by nano-UHPLC−MS/MS.35,40 N-terminal enrichment strategies have not been applied to mycobacterial proteins or secreted protein populations.
We developed a novel one-pot filter-aided sample prep (FASP) for quantitative N-terminal enrichment, followed by mass spectrometry. We applied this approach to the cytosolic and secreted proteomes of both M. marinum and M. tb. We present the first large-scale quantitative and comparative study of NTA in Mycobacterium. We partially validated by comparing them against the large-scale mycobacterial proteome of Kelkar et al.30 Using our data, combined with published literature, we generated commonly found amino acids at protein N-termini for NTA and functional classification of NTA mycobacterial proteins. We defined the ratios of partially acetylated proteins across cellular compartments linking differential acetylation to protein localization. Our findings reveal novel biology underlying the NTA of mycobacterial proteins that may provide a basis to understand NTA in mycobacterial physiology, pathogenesis, and antimicrobial resistance.
EXPERIMENTAL PROCEDURES
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Growth of Mycobacterium
M. marinum M strain was acquired from ATCC (BAA-535) and maintained in at 30 °C in Middlebrook 7H9 broth (Sigma-Aldrich, St. Louis, MO) with 0.5% glycerol and 0.1% Tween-80 as previously described.61 M. tb Erdman (Gift of Jeffery S. Cox) was maintained in 7H9 with the addition of 10% oleic albumin dextrose catalase (OADC, BD Biosciences), 0.5% Tween-80, and 0.5% glycerol and grown at 37 °C as previously described.35
Generation of Mycobacterial Protein Fractions
Secreted and cell-associated protein fractions from Mycobacterium were prepared as previously described.35,61 In brief, M. marinum and M. tb were grown under conditions permissive for ESX-1-mediated protein secretion. Following growth, bacterial cells were harvested by centrifugation. Culture supernatants were isolated by filtration and concentrated. Protein concentrations were determined using the MicroBCA kit (Pierce) according to the manufacturer’s instructions.
Synthesis of N-Acetoxy-D3-succinamide/N-Acetoxy-succinamide
N-Acetoxy-D3/H3-succinamide was synthesized as described by Staes et al.57 In brief, 1 g of D6 acetic anhydride (9 mmol) was incubated with 350 mg (3 mmol) of N-hydroxysuccinimide with stirring overnight. The reaction was then stirred for an additional 8 h uncovered, purified, and dried by rotary vacuum and six exchanges with 3 mL of n-hexanes (Sigma-Aldrich). After desiccation, activity was measured by alkylation of leu-enkephalin (YGGFL) and measured on a MALDI-MS (data not shown). Purity was estimated at >95% by mass.
Filter-Aided Sample Preparation Compatible Protein Digestion
500 μg of M. marinum and M. tb protein preparations (cell associated and secreted) were acetone or methanol−chloroform precipitated as previously described.62 N-terminal enrichment was performed using a protocol modified from Staes et al.57 Precipitated proteins were resuspended and solubilized with a 1:2 ratio of 0.4% SDS and 100 mM DTT in 100 mM triethylammonium bicarbonate (TEAB), incubated at 95 °C for 5 min, and cooled for 3 min on ice. The denatured and reduced protein fractions were loaded onto a 10 000 molecular weight cutoff (MWCO) Amicon filter (Millipore, Billerica, MA) and washed twice by the addition of 200 μL of 8 M urea in 100 mM TEAB with centrifugation. Cysteines were alkylated by adding 200 μL of 50 mM iodoacetamide in urea/TEAB for 5 min in the dark, followed by centrifugation. Free primary amines, lysines, and unblocked N-termini were acetylated directly in the concentrator by incubation of the prepared proteins with 10 mM N-acetoxy-D3-succinamide (final) dissolved in acetonitrile (ACN), then diluted (1:1) with 500 mM TEAB/5% isopropanol at room temperature for 1 h with gentle rotation. An additional 10 mM N-acetoxy-D3-succinamide was added for a second 1 h of incubation at room temperature with gentle rotation. The acetylation reaction was quenched by the addition of 40 μL of 1 M glycine for 10 min and then treated with 40 mM hydroxylamine for 10 min at 30 °C, followed by two spin-washes with 50 mM ammonium bicarbonate. Trypsin or GluC (Promega, Madison, WI) was added at a protease to substrate ratio of 1:100, and digestion proceeded overnight at 37 °C. Trypsin cleaves after Lys and Arg. GluC cleaves after Glu under these conditions. Digestions were terminated with phenyl−methyl sulfonyl fluoride (PMSF) to a final concentration of 1 mM and allowed to stand for 60 min at room temperature to inhibit the protease and eliminate the PMSF from the solution through hydrolysis.63
Removal of Pyroglutamate N-Termini
Acidic peptide termini can chemically modify into N-terminal pyroglutamic acid following digestion. Neo N-terminal Glu and Gln residues cyclize, forming a lactam. Pyro-Glu and Gln → Pyro-Glu (via deamination) therefore reduce the charge on the N-terminus of a peptide, which results in elution on cation-exchange, comingling with protein N-termini. To reduce this effect, cyclized amino acids were removed using the pGAPase/Qcyclase treatment as described in Staes et al.57 pGAPase was purified using the Tagzyme kit (Qiagen, Valencia, CA57), and 625 mU was activated by adding 1 μL of 800 mM NaCl, 1 μL of 500 mM EDTA, and freshly prepared cysteamine to a final concentration of 3 mM and incubating at 37 °C for 10 min. Qcyclase (1250 mU) and activated pGAPase were then added to the filtration concentrator and incubated at 37 °C for 1 h. Peptides were subsequently eluted from the filtration concentrator by centrifugation with a second extraction using ~150 μL of 500 mM NaCl. Samples were desalted using a C18 50 mg solid-phase extraction (SPE) cartridge (Waters, Milford, MA) and dried in a Speed Vac.
Fractionation of Peptides for N-Terminal Peptide Enrichment
For trypsin and GluC derived peptides, strong-cation exchange (SCX) and strong-anion exchange (SAX) were employed, respectively.
SCX.
Peptides were resuspended in 0.5 mL of 0.08% TFA at pH 3.0 and then diluted 1:1 with acetonitrile. Peptides were fractionated on a 50 mg SCX SPE (Phenomenex). The column was equilibrated with three column volumes of 10 mM NaPO4 pH 3.0 in 50% ACN. The sample was loaded onto the column, and the flow-through fraction was collected. Additional washes with 10 mM NaPO4 pH 3.0 in 50% ACN were collected, resulting in a final volume of 5 mL. The column was eluted using three column volumes of 5% NH4OH. Flow-through fractions (containing enriched N-termini) were desalted using C18 spin columns per the manufacturer’s instructions (Protea, Morgantown, WV). Eluted fractions were desalted using C18 1 mL−50 mg SPE columns. All samples were then prepared for LC−MS/MS analysis.
SAX.
Peptides were resuspended in 1 mL of 30% ACN and 20 mM Tris pH 8.0. The column was equilibrated with the same solution using three column volumes, and the sample was loaded onto the column. The column was washed with the same solution, and flow-through fractions were collected. Peptides were eluted using stepwise salt additions of three column volumes of 20, 100, and 500 mM NaCl in 20 mM Tris pH 8.0 to collect N-terminal enriched fractions. Fractions were desalted using C18 spin columns according to the manufacturer’s instructions (Protea, Morgantown, WV) and then dried prior to LC−MS/MS analysis.
Mass Spectrometry
Desalted digests of enriched and depleted fractions were analyzed by bottom-up LC−MS/MS essentially, as described in Williams et al.61 In brief, peptides corresponding to ~1 μg of digest were loaded onto C18 BEH column 100 μm × 100 mm (Waters, Billerica, MA) and separated over a 90−120 min gradient from 5 to 35% aqueous (A) to organic (B) phase (A = H2O + 0.1% formic acid; B = acetonitrile + 0.1% formic acid) running at 800 nL/min. The column outlet was directly connected to an electrospray emitter, and MS and MS/MS data were acquired on an LTQ Velos Orbitrap FTMS instrument (Thermo, San Jose, CA) (three technical replicates, first biological replicate) or Q-Exactive HF FTMS instrument (three technical replicates, second biological replicate). The LTQ Velos Orbitrap acquired a TOP12 data-dependent acquisition (DDA); the Q-Exactive was acquired in TOP18 DDA mode.
Data Analysis
RAW data files were converted to Mascot Generic Format (.mgf) peak lists using Proteome Discoverer v 1.4 (Thermo). mgf peak lists were searched using Protein Pilot running the Paragon algorithm (V5.0) in Rapid and Thorough mode with trideuteroacetylation “predigestion” and acetylation emphasis as a special factor.64 FASTA files of M. marinum and M. tb were downloaded from MarinoList (release 22/2010) and Tuber-culist (release 2.6/2013) as of 01–2018.65 A list of common contaminants was appended to each FASTA prior to search (concatenated FASTA = 13 728 proteins 27 456 w/decoy). False discovery rates (FDRs) were determined using the target-decoy method of Elias et al.66 Cutoff for all data interpreted was at an FDR = 0.01 or better. Processed and RAW data are available at MassIVE at accession number MSV000081168 (ftp://massive.ucsd.edu/MSV000081168).
Bioinformatics
Peptides used for N-terminal analysis were identified by sorting by modification and comparing N-termini manually to the canonical N-terminus of the respective protein. N-termini spectra were inspected using Qual Browser (Thermo). IceLogo, an open-source software for the analysis and visualization of consensus patterns in peptide sequences, was used to analyze N-terminal motifs (http://iomics.ugent.be/icelogoserver/index.html).67 The N-terminal sampling method with a p-value cutoff of 0.05 was used. This method compared the first six amino acids in context to the canonical proteome. For the reference sets, UniProt (uniprot.org) canonical proteomes for the respective organisms were obtained and uploaded to IceLogo. Venn diagrams were calculated using the Venn Diagram Generator from the Whitehead Institute (http://jura.wi.mit.edu/bioc/tools/venn.php).
Experimental Design and Statistical Rationale
For M. tb, three biological replicates were analyzed in technical duplicate. One additional data set had poor overall spectral density and was discarded, although it was “searched”. For M. marinum, three biological replicates were analyzed in technical duplicate. All data sets were filtered at a 1% FDR as per the method of Elias et al.66 Replicate data sets for samples digested with trypsin or GluC were combined for each search (by species of mycobacteria), generating an aggregate FDR of 1%. This approach reduces cumulative false discovery events for low-frequency peptide hits with less spectral evidence (high local FDR %). Peptides fewer than six amino acids were not considered for N-terminal peptide analysis. Because of quantitative enrichment, spectral redundancy is high (Table 1). We also subjected the “discarded” neo-peptide fractions to LC−MS/MS analysis because they contain unenriched N-termini, and the remaining tryptic peptides provide additional statistical weight to confirm peptide spectral match (PSM) for proteins. Percent acetylation was performed as described below; error was calculated as the average of the protein error determined by label-free-quantification (LFQ)61 plus a correction for average isotope overlap from heavy into light, which we averaged at 10%. This was determined using the modal peptide length (~12 aa) and calculating the isotopic contribution for Averagine (C4.94H7.76N1⋅36O1.15S0.04). For large peptides >18 aa, we integrated the first isotope peak for light and heavy to minimize this contribution to <5%.
Table 1.
N-Terminal Acetylation in Mycobacteria
| analysis by protein ID | M. tb Erdman | M. marinum M |
|---|---|---|
| detected proteome | 1910 | 2375 |
| total NT IDsa | 498 | 838 |
| acetylation at P1’/P2’ | 211 | 347 |
| iMet/P1’ | 50 | 88 |
| P2’ | 169 | 272 |
| heavy acetylation of P1’/P2’ | 369 | 665 |
| iMet/P1’ | 79 | 163 |
| P2’ | 303 | 537 |
| partial acetylationb | 82 | 174 |
| frequency at residue T/M/S/Ac | 27/22/19/21 | 39/24/16/10 |
| fMet | 11 | 16 |
Numbers in this Table are representative of nonredundant protein ID (e.g., only a single proteoform is counted per polypeptide) for each category.
Polypeptide found both N-terminally acetylated and heavy acetylated.
Percentage of NT-acetylated peptides with NTA events at Thr, Met, Ser, and Ala residues. The remaining NTA events that occurred at other residues are lower frequency and can be found in Supplemental Tables 1 and 3.
RESULTS AND DISCUSSION
Development of a Filter-Aided Sample Preparation Compatible Approach To Enrich and Quantify NTA
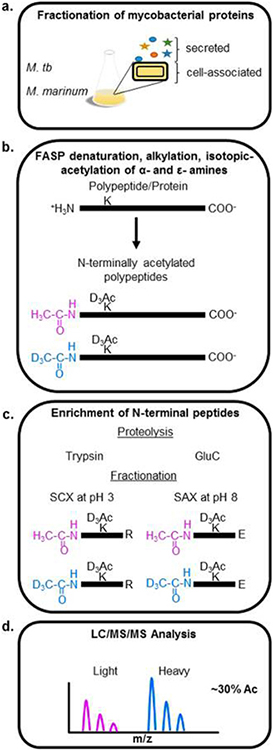
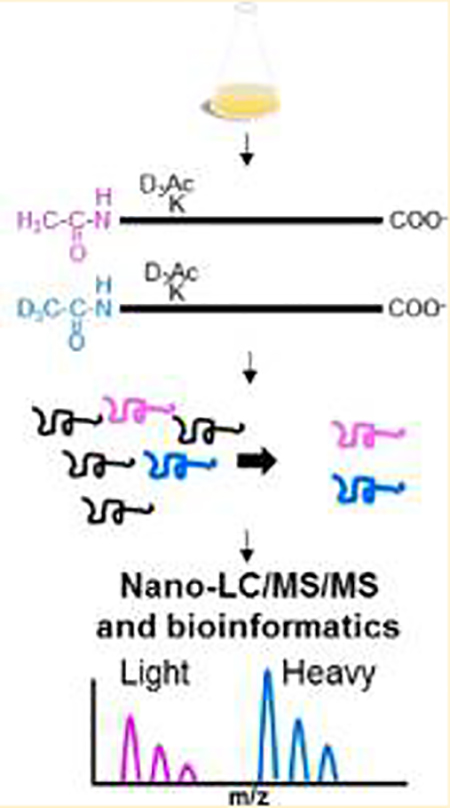
We sought to enrich and quantify the modification of mycobacterial protein N-termini as a necessary first step in understanding NTA in mycobacteria. Although powerful approaches exist to interrogate large swaths of a proteome on an LC−MS/MS time scale, these methods favor protein identification, rather than specific detection of a type of polypeptide, like protein N-termini.35 We developed a simplified workflow for the enrichment and quantification of mycobacterial protein N-termini (Figure 1) by adapting existing N-terminal enrichment protocols.57 We condensed the enrichment of protein N-termini to a single filter-aided sample preparation (FASP) method. Samples were prepared for MS analysis, as outlined in Figure 1. Because the majority of reported NT-acetylated mycobacterial proteins are secreted virulence factors, we analyzed both secreted and cell-associated protein fractions from M. tb Erdman and M. marinum M for NTA (Figure 1a). The protein fractions were denatured and reduced, then alkylated in a 10 000 MWCO filter unit. Primary amines on unacetylated protein N-termini and Lys side chains were acetylated with an N-acetoxy-D3-succinamide (D3Ac, Figure 1b). This step allowed us to distinguish between chemical and in vivo acetylation (i.e., “exogenous” vs “endogenous” labeling). Acetylated Lys residues prevent cleavage by trypsin, which reduces the number of tryptic sites at the N-terminus, improving coverage.
Figure 1.
Mycobacterial protein N-terminal enrichment. (a) Triplicate cultures of M. tb and M. marinum grown in Sauton’s minimal media were fractionated into cell-associated (cytosolic) and secreted fractions. (b) Protein fractions were denatured and transferred to a 10 kDa centrifugal concentrator, alkylated, and stable-heavy isotope-acetylated at α- and ε-amines with N-acetoxy-D3-succinamide. (c) Proteins were digested with trypsin or GluC proteases, and the resulting peptides were fractionated using SCX or SAX, respectively, to enrich for N-terminal peptides. At pH 3, N-terminal peptides are net-neutral charge and will preferentially flow through SCX extraction. At pH 8, N-terminal peptides digested with GluC possess an extra carboxy group at the C-terminus and will have a net-negative charge, which can be enriched by binding to a SAX column. (d) N-terminally enriched fractions were analyzed by at least triplicate nano-UHPLC−MS/MS, and the extent of acetylation was determined using the ratio of stable “heavy” isotope acetyl to that of the endogenous “light” acetyl group on duplicate samples (2 M. marinum, 2 M. tb).
The choice of enzymes limits the scope of most global and enriched proteomes. Trypsin is the preferred protease for the majority of bottom-up proteomics due to its high kcat, specificity, and suitability for mass spectrometry. In silico analysis of the canonical M. tb Erdman proteome revealed that 48% of proteins contain an Arg residue within the first 11 aa (Figure S-1). The abundance of Arg residues at the N-terminus of mycobacterial proteins produces small N-terminal peptides following digestion with trypsin that are difficult to correlate to a single protein identification.35,68,69 Published bacterial NTA studies do not address this problem.15–19 We reasoned that using a separate protease with different specificity from trypsin would promote the resolution of additional protein N-termini.68 To this end, we screened several proteases in silico for their cleavage specificity at mycobacterial protein N-termini. We determined that GluC protease, which cleaves after Glu and Asp residues, could allow detection of an additional 30% of the canonical N-terminal proteome unobservable from a trypsin digest alone. Therefore, the blocked proteins were digested with trypsin or GluC proteases separately to maximize representation of the N-terminal mycobacterial proteome. GluC also has the advantage of minimizing N-terminal cyclization (e.g., pyro-glutamic acid formation), potentially reducing the necessity for pGAPase/Qcyclase treatment. GluC cleaves C-terminal to Glu and Asp residues. Glu residues will not be present at exposed N-termini and therefore cannot cyclize. Pyro-glutamic formation remains a concern for Gln residues.
The digested peptides were desalted and termini-enriched on SCX or SAX solid-phase extraction for trypsin or GluC digests, respectively (Figure 1c). SCX and SAX separate neopeptides (+NH3) from blocked peptides (i.e., all N-termini). Because of improvements in instrument performance and the relatively simplified proteomes of microbes, enriched peptide fractions were subjected to a single dimension of nano-UHPLC−MS/MS, identified by target-decoy database search, and aligned.70 “Light” (H3Ac) to “Heavy” (D3Ac) ratios of N-terminally labeled peptides were quantified to derive the proportion of endogenous protein acetylation (Figure 1d).
Enrichment of Protein N-Termini in M. tb
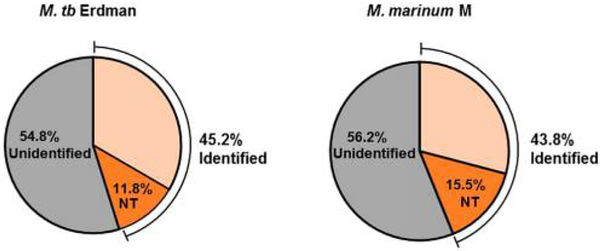
We analyzed our data to determine the representation of the mycobacterial proteomes and the enrichment for protein N-termini in our resulting fractions. Our findings are presented as Table 1 and Figure 2. We identified 1910 proteins from M. tb Erdman (1% FDR, Table 1), which corresponds to 45.2% of the canonical proteome (Figure 2, left). Of the detected M. tb proteome, 498 nonredundant protein identifications (IDs) were made from N-terminal peptides (N-terminal peptide ID, Table 1). On the basis of these data, we detected at least 11.8% of the canonical N-termini in M. tb (Figure 2, left). 26% of identified proteins had assignable termini. N-termini comprised 25% of the enriched peptide fractions in SCX as compared to 4.8% in the unenriched peptide fraction. For SAX, N-termini constituted 11.8% of the enriched peptide fractions as compared with 4% in the unenriched peptide fraction. From these data, we conclude that we successfully developed a one pot protocol to positively enrich protein N-termini from M. tb proteomes.
Figure 2.
Graphical distribution of detected proteins by LC−MS/MS in M. tb and M. marinum. Protein identifications in this study for (a)M. tb Erdman (left) and (b) M. marinum M (right). We identified45.2 and 43.8% of the canonical proteomes for each species. All of the proteins included internal or neopeptides for the identification. 11.8 and 15.5%, respectively, were also identified using their N-terminal peptides.
We identified 211 proteins from M. tb that were endogenously N-terminally acetylated (Table 1, Supplemental Table 1). Thus 42.3% of the observed N-terminal proteome (211/498) and 11% of the observed total proteome (211/1910 proteins) were N-terminally acetylated under the conditions tested for M. tb. Acetylation of mycobacterial proteins was measured on both the initiator methionine (iMet) and the second residue generated following iMet cleavage (Table 1). The N-terminally acetylated proteins, either on the iMet or on the second residue, are listed in Supplemental Table 1. From these data, we conclude that our protocol successfully enriched and identified N-terminally acetylated proteins from M. tb.
To validate our enrichment strategy, we compared our results with the proteome of M. tb H37Rv by Kelkar et al.30 (Figure S-2, Supplemental Table 2). They employed a shotgun proteogenomic approach and did not specifically enrich proteotypic N-termini. We extracted 247 proteins with canonical NTA from the Kelkar et al. data set.30 We detected 86 of these proteins in our study. 122 of the proteins we identified were not found in the whole-proteome approach. Thus neither approach has exhaustively identified NTA in M. tb. However, additional differences between our study and Kelkar et al. may have also contributed to differences in the detected NTA proteins. Specifically, the strain background and the growth conditions differed between the two studies. First, although both Erdman and H37Rv are widely used laboratory strains, Erdman is considered more virulent than H37Rv in relevant infection models. Second, we grew M. tb in Sauton’s media, under secretion permissive conditions, while Kelkar et al. grew M. tb in 7H9 for a defined period of time. Because we do not know the growth phase of the bacteria in both studies, we cannot rule out media or growth phase as factors contributing to the different NTA identifications. Together, our study and our analysis of the published findings from Kelkar et al.30 identified 368 N-terminally acetylated proteins in M. tb (Figure S-2, Supplemental Table 2).
Comparative Analysis of NTA in M. tb and M. marinum
We identified 2375 proteins from M. marinum (1% FDR, Table 1), which corresponds to 43.8% of the canonical proteome (Figure 2, right). 838 nonredundant protein identifications were made from N-terminal peptides (NT Peptide IDs, Table 1). We detected 15.5% of the canonical M. marinum protein N-termini. As in M. tb, we conclude that our method was successful in enriching protein N-termini from the M. marinum proteome (Figure 2).
Of the detected N-terminal peptides, 347 peptides from M. marinum were endogenously N-terminally acetylated (Table 1, Supplemental Table 3). Thus, of the observed N-terminal proteome, 41.4% of detected N-termini (347/838) or 14.6% of the observed proteome (347/2375 proteins) were N-terminally acetylated in M. marinum under the conditions tested. As with M. tb, acetylation of M. marinum proteins was observed on both the iMet and the second residue following iMet cleavage (Table 1). The N-terminally acetylated proteins, either on the iMet or on the second residue, are listed in Supplemental Table 3 for M. marinum. From these data, we conclude that NTA of M. marinum proteins is similar in scope to M. tb.
To our knowledge, our study reports the first targeted enrichment of protein N-termini in mycobacteria. While several studies have focused on N-terminal enrichment for identifying NTA in bacteria,16,17 our approach is simpler and can be performed with modest investments in instrument time. We enriched termini approximately five-fold (by frequency) relative to the detected proteome (~90% by mass enrichment) with relatively low amounts of samples and instrument use. Moreover, our approach couples the high-efficiency digestion provided by spin-filter proteomics (FASP) with biochemical enrichment of protein termini. Here we dramatically decreased sample handling, cleanup, and fractionation steps, enabling a lysate-to-N-terminome generation in about 1 day. Moreover, using GluC in parallel with trypsin allowed us to observe an additional 5% of the total proteome that was not accessible with trypsin digestion. This represents 22% more termini identified using GluC with 18% additional NTAs identified. The use of multiple proteases increases the probability of identifying protein N-termini and is likely essential when using bottom-up proteomic approaches for N-terminal enrichment studies.
N-Terminally Acetylated Mycobacterial Proteins Are Functionally Diverse
We conducted functional analyses of the N-terminally acetylated protein populations we identified to gain insight into the role of NTA in mycobacteria. Functional annotation of the M. tb Erdman proteome is incomplete as compared with the more widely used laboratory strain, M. tb H37Rv (UniProt, uniprot.org; BioCyc, biocyc.org).71 Accordingly, we used the corresponding M. tb H37Rv orthologs of the NT-acetylated proteins we identified in the Erdman proteome (Supplemental Table 1). Of the 211 N-terminally acetylated proteins we identified from M. tb, 208 (98.5%) had clear orthologs in the H37Rv strain.
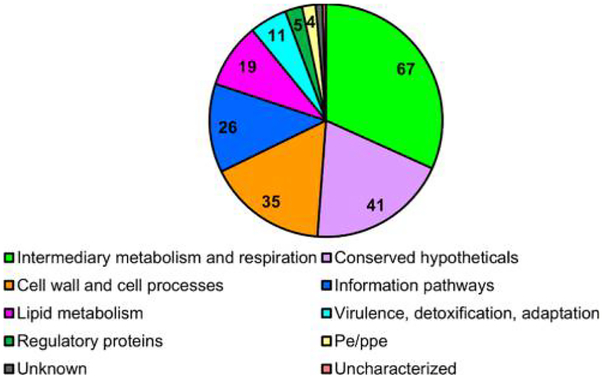
We defined the functional categories of the M. tb N-terminally acetylated proteins based on the annotations listed in Mycobrowser (Figure 3, Supplemental Table 1). 67 of the NT-acetylated proteins (31.8%) were categorized as intermediary metabolism and respiration (TCA). The large percentage of NT-acetylated proteins categorized as central metabolism is consistent with the scope of protein NTA identified in bacteria to date.16,17 We observed similar trends in M. marinum (Figure S-3). 41 were classified as conserved hypotheticals, 35 as cell wall and cell processes, 26 as information pathways, 19 as lipid metabolism, 11 as virulence, detoxification, and adaptation, 5 as regulatory proteins, and 4 as PE/PPE proteins, and 3 were not assigned a category. In addition to those characterized as virulence, detoxification, and adaptation, many proteins we identified, including the PE/PPE proteins and several of those characterized as cell wall and cell wall processes, are also necessary for virulence.72–75 Together, our findings indicate that virulence proteins and those involved in metabolism likely represent the two largest categories of acetylated proteins we detected in pathogenic mycobacteria.
Figure 3.
Functional characterization of M. tb Erdman N-terminally acetylated proteins. Mycobrowser65 was used to identify the functional categories of the N-terminally acetylated proteins identified in this study. Two proteins were classified as “unknown” and one protein was classified as “uncharacterized”.
Our findings demonstrate that NTA spans broad functional distributions of mycobacterial proteins. Our measurements may be biased due to the generally high abundance of proteins required for growth.76 Indeed, proteins required for growth and protein synthesis are thought to have long half-lives in cells and NTA may promote protein stability.6,7 However, we propose that our findings likely reflect mycobacterial physiology. Consistent with previous reports of NTA, we identified a substantial number of secreted NT-acetylated virulence factors associated with the ESX-secretion systems.34,36–38 Our findings are consistent with functional analyses conducted on NT-acetylated proteomes from Pseudomonas and Acinetobacter, in which NT-acetylated proteins were involved in transcription, translation, amino acid biosynthesis, and central metabolism and pathogenesis.16,17
Specific Amino Acids Are Enriched at Acetylated Protein N-Termini
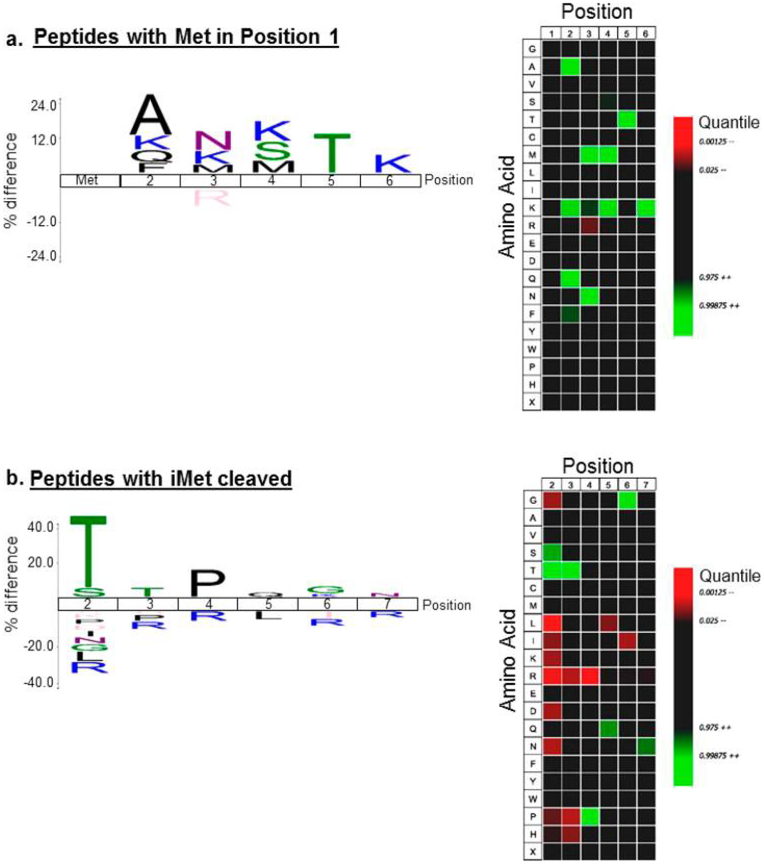
NATs catalyze NTA. The first six amino acids are sufficient for NAT activity in vitro,10,77 indicating that substrate specificity may be prior to protein folding. We used IceLogo67 analysis of the NT-acetylated peptides enriched from M. tb Erdman relative to the M. tb Erdman reference proteome (UniProt, UP000007568) to define if the first six amino acids contained a conserved motif for specificity of NTA. We first analyzed the 50 unique peptides that were NT-acetylated on the iMet residue (P1′ position). As shown in Figure 4a, the most common amino acids following the iMet can be summarized as follows: [A/K/Q/F]-[N/K/M]-[K/S/M]-[T]-[K]. Ala, Lys, and Gln were enriched at the P2′ position. Gln, Lys, and Met were enriched at the P3′ position. Conversely, Arg was significantly underrepresented at the P3′ position. Lys, Ser, and Met were enriched at the P4′ position. At P5′, Thr was enriched. Interestingly, Lys was specifically enriched at several positions in iMet containing acetylated peptides. Trypsin does not proteolyze Lys residues under these conditions due to the trideuteroacetylation of Lys prior to proteolysis. Therefore, the observed enrichment of Lys in the iMet containing peptides was not an artifact of the preparation. We did not observe Lys enrichment on NTA peptides in which iMet was cleaved (see below).
Figure 4.
IceLogo analysis of N-terminally acetylated protein populations for M. tb. Peptides identified with endogenous NTA were analyzed against the canonical M. tb proteome using IceLogo. IceLogos (left) and Heatmaps (right) are shown for (a) 50 N-terminally acetylated peptides with intact iMet and (b) 169 N-terminally acetylated peptides with cleaved iMet. For the IceLogos and the Heatmaps, only significant (P < 0.05) amino acids are shown. For Heatmaps, green denotes enrichment and red denotes underrepresentation of amino acids at the designated polypeptide position.
We analyzed the 169 unique peptides lacking the iMet and that were NT-acetylated on the P2′ residue (Figure 4b). We found that peptides acetylated on the P2′ residue were almost exclusively acetylated on Thr as compared with sampling from the canonical N-terminal proteome. Ser was the second most frequently acetylated residue. The most common amino acids in the first six positions for NT-acetylated M. tb Erdman peptides (at P2′) can be summarized as [T/S]-[T]-[P]-[Q]-[G/K]-[N] (Figure 4). We found that specific amino acids were also underrepresented at the P2′ position (His, Pro, Asp, Ile, Asn, Gly, Leu, and Arg). This is likely due in part to the N-end rule in which Leu, Lys, and Arg promote protein degradation in bacteria.78 There was an absence of Arg at the P3′, P4′, and P6′ residues as compared with the reference proteome. Arg and Ile were underrepresented at the P5′ position. Similar analysis of the NT-acetylated peptides from M. marinum revealed that the most common amino acids in the first six positions were [T]-[T/S/E]-[P/A]-[E/N/D]-[E/T/S/N]-[P/D] (Figure S-4). Searching for these patterns against the canonical M. tb and M. marinum proteomes using Mycobrowser revealed that these particular combinations of amino acids were not found together in the NTA proteins that we identified, suggesting that these amino acids do not likely make up a motif for NTA. Our data are not dense enough to determine if the differences between M. tb and M. marinum are significant. In NTA peptides from both species, Thr was enriched at P2′ and Pro was enriched at P3′. In all cases, enrichment against Arg is likely a consequence of the cleavage with trypsin as it would generate short two- to six-amino-acid peptides, which are not identifiable in most experiments.
Finally, we determined the amino acids most commonly found in the first six positions of protein N-termini that were chemically heavy-isotope-acetylated (native unacetylated N-termini, from Figure 1 and Table 1) in M. tb and M. marinum to rule out biases in our methodology (Figures S-5 and S-6). The amino acids enriched at the P2′−P6′ residues in this population were distinct from those discovered for the endogenously acetylated peptides. The chemically acetylated peptides were enriched for charged residues within the first six amino acids, similar to peptides acetylated on the iMet (Figure 4b). The Pro at position P3′ was conserved only among endogenously acetylated peptides. From these data, we conclude that the observed enriched residues at the N-termini of endogenously acetylated proteins from M. tb and M. marinum were not due to bias in the assignment of NTA but rather reflect differences in the NT-acetylated protein populations.
Noncanonical protein N-termini are termini that are not at the predicted start site of the protein. NTA of noncanonical N-termini represent potential sites of post-translational modification. We analyzed the NTA of noncanonical peptides in M. tb, excluding those peptides with N-terminal trypsin or GluC cleavage sites. 186 nonredundant peptides corresponding to 147 M. tb proteins were analyzed using IceLogo with a random analysis against the canonical M. tb Erdman proteome (Figure S-7, Supplemental Table 4). Interestingly, residues enriched in canonical NTA profiles for M. tb and M. marinum were also enriched, although not exclusively, in the noncanonical N-terminally acetylated population and can be summarized as [T/S/P/M]-[A/T]-[A/K/P]-[K]-[E/K/H/M]-[A/K].
Our findings demonstrate that the mycobacterial proteins have unique signatures for NTA. Bienvenut et al. found that in E. coli proteins with processed Met targeted for NTA at Ser commonly contained Glu at P4′ and Lys at P6′.15 Kentache et al. found that in A. baummanii Met-processed N-termini acetylated at Asn contained basic residues at P6′−P9′.17 M. marinum proteins are NT-acetylated at Thr residues almost exclusively, which is in sharp contrast with other protein acetylation studies, where Ala and Ser were enriched for NTA.4,5,16 The M. tb and M. marinum genomes include 27 and 31 genes (24 that are shared) predicted to encode for NATs. Therefore, although we identified a conserved signature of commonly found amino acids in the first six positions of NT-acetylated peptides, we cannot determine if this region confers specificity to a particular NAT. Our findings and those from previously published reports need to be followed up to determine if the identified patterns are sufficient or necessary for NTA of bacterial proteins in vivo and to link NATs to their targets.
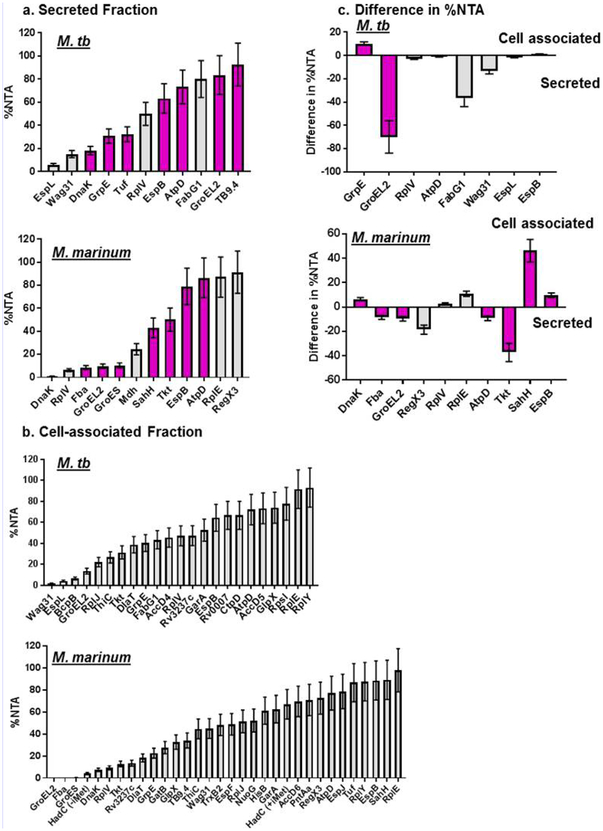
Quantification of Acetylation in Cell-Associated and Secreted Protein Fractions
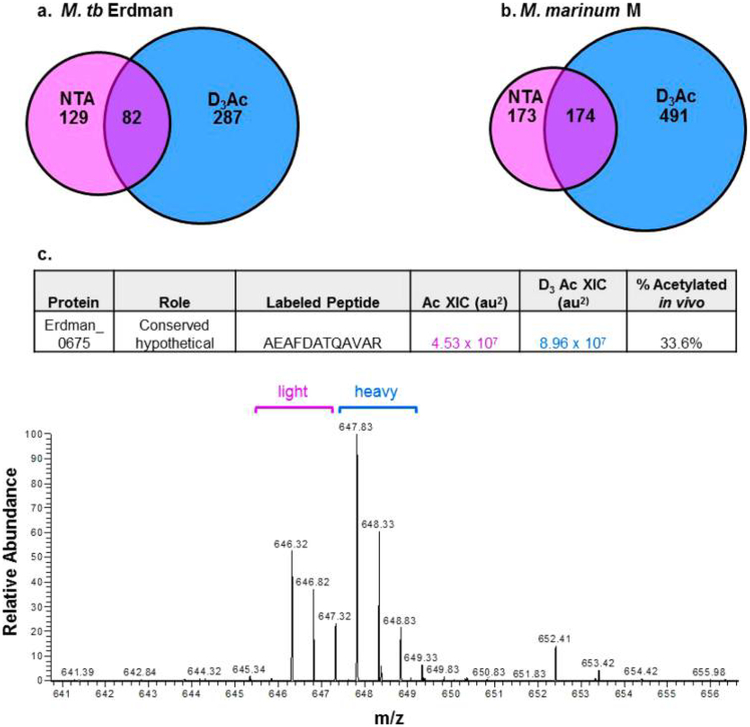
To advance our understanding of NTA, we quantified the levels of NTA on mycobacterial proteins that were cell-associated or secreted from the cell. We labeled intact unmodified protein N-termini with stable heavy-isotope acetyl groups to distinguish protein termini from the background of neo-peptides. Of the 211 and 347 endogenously acetylated proteins identified in this study (Table 1), 82 and 174 proteins from M. tb (Figure 5a) and M. marinum (Figure 5b), respectively, were also labeled with N-acetoxy-D3-succinamide. Acetylation of intact proteins with N-acetoxy-D3-succinamide increases the mass of the N-terminally acetylated peptide by three (Da) as compared with an endogenously acetylated peptide (45 vs 42 nominal mass Δ). In this way, we could calculate the relative ratios of acetylation by identifying the light and heavy m/z peaks for unique peptide fragment pairs (Ac and D3Ac). The peaks were integrated as extracted ion chromatograms (±3 ppm), and the area was determined with a five-point Gaussian-smooth. The percentage of acetylation (% NTA) for each unique protein N-terminus was determined by the ratio of [Ac−H3]/([Ac−H3] + [Ac-D3]) in duplicate (representative spectra, Figure 5a). The %NTA provided an inference of the proportion of NT-acetylated protein in both cell-associated and secreted protein fractions. Error was determined as the average error of the protein, as determined by LFQ (Figure 6), plus a percentage determined as the average contribution from isotope overlap of 10%, as previously described.
Figure 5.
Quantifying N-terminal acetylation. Distribution of N-terminal identifications in (a) M. tb and (b) M. marinum. Magenta denotes NTA protein identifications, blue denotes D3Ac (exogenously acetylated) protein identifications, and the intersections are protein IDs found in both forms and that can be targeted for relative quantitation. (c) Example spectrum generated by LC−MS/MS analysis of peptide pairs targeted for acetylation quantitation. 646.32 m/z is the endogenously acetylated Erdman_0675 peptide (magenta bracket, “light”). 647.83 m/z corresponds to the D3 chemically acetylated peptide (blue bracket, “heavy”). Peaks were integrated by extracted ion chromatogram ±3 ppm, and the percentage of acetylation was calculated using % Ac in vitro: (Ac area)/(Ac area + exogenously Ac area).
Figure 6.
Measurement of NTA ratio in cell-associated and secreted fractions. M. tb and M. marinum measurements of NTA for H37Rv protein orthologs in (a) secreted and (b) cell-associated fractions. Measurements were taken from single points. (c) Difference in %NTA of selected peptides between the cell-associated and secreted fractions. The difference was calculated by subtracting the %NTA in the secreted fractions from the %NTA in the cell-associated fraction. Error bars represent the average error of the protein in LFQ data set (±20%). For panels a and c, the magenta bars represent M. tb H37Rv proteins that have been identified in the secreted fraction in previous studies as reported by Mycobrowser.
We next determined if the acetylation status of orthologs among M. tb and M. marinum were conserved. We identified 72 M. tb H37Rv shared orthologs in our data set among the acetylated populations. Of these, 26 and 33 N-terminal peptide pairs for M. tb and M. marinum were suitable for quantification. Sixteen had shared protein orthologs in the cell-associated fraction and five were shared in the secreted fraction. The peptides used for quantification in the cytosolic and secreted protein fractions are listed as Supplemental Tables 5 and 6.
Interestingly, the NTA of the majority of M. tb and M. marinum proteins was not 100%. Instead, the mean NTA of the M. tb and M. marinum proteins we quantified was 49 and 44%, ±20%, respectively (Figure 6). Moreover, a fraction of the proteome contained at least four separate protein species: ± N-terminal methionine and ± NTA. These findings indicate that partial acetylation may be the most frequent form of acetylation in Mycobacterium. Whereas we do not know the mechanism underlying partial NTA, it is possible that the N-terminal motif dictates the level of NTA. Alternatively, NTA could affect protein stability or localization, which could impact the total level of NTA.
We found that proteins involved in information pathways such as the 50S ribosomal subunits RplE and RplY were almost fully N-terminally acetylated in the cytosol in both M. tb and M. marinum. This finding is consistent with a model in which NTA promotes protein stability for housekeeping proteins.79
We observed differential acetylation in the cell-associated versus secreted fractions for specific M. tb and M. marinum proteins (Supplemental Table 7 and Figure 6). In M. tb, GroEL2 was found in the Met-processed form in the cytosolic and supernatant fractions at 14 and 84%, respectively. Conversely, in M. marinum, GroEL2 was unacetylated in the cytosolic fraction and 10% acetylated in the secreted fraction.
Interestingly, the level of acetylation of the ESX-1 substrate, EspB, was high in M. tb and M. marinum. EspB was 60−90% acetylated in both the cytosolic and secreted protein fractions (Supplemental Table 7 and Figure 6). We previously observed the acetylation status of secreted EspB to be 76.65%.34 The N-terminal sequences of EspB in M. tb and M. marinum are M.TQSQTV… and M.SQPQTV…, respectively, both of which resemble the sequences we identified for NTA in this study.
Some of the proteins identified in the secreted fraction are likely contaminants from low-level lysis, as they are not known to be secreted. One such protein, AtpD, was acetylated at comparable ratios when found inside and outside of the cell in M. tb and M. marinum. RplV is a ribosomal protein that was also found in both the cytosolic and supernatant fractions in M. tb and M. marinum. RplV was 47 to 48% acetylated in M. tb and 6−9% acetylated in M. marinum (Supplemental Table 7 and Figure 6). The levels of acetylation in the cytosolic and secreted fractions indicate that RplV acetylation is low compared with other ribosomal proteins, indicating a potential point of regulation. RplV in E. coli binds the 23S rRNA of the 50S subunit and is stimulated by the ribosomal proteins L4, L17, and L20.80 If and how acetylation impacts the assembly of the 50S ribosomal subunit would be an interesting avenue for future study. These proteins may serve as future lysis controls for NTA quantification because they remain consistent in acetylation ratio among cytosolic and secreted fraction.35,36
To our knowledge, this is the first targeted quantification of protein N-termini in mycobacteria. Together, our findings indicate that N-terminal peptides from M. tb and M. marinum proteins exhibit specific NTA levels, which can vary based on the presence of the iMet or the biological localization (cell-associated vs secreted). We suspect that other proteins N-terminally acetylated in the presence and absence of iMet exist in the proteome but were not quantifiable with our cutoffs. Our findings that protein NTA is linked to localization suggest a regulatory mechanism for protein transit across the mycobacterial cell wall. Indeed, our previous findings suggest that NTA may promote the transport of EsxA, an ESX-1 substrate, across the mycobacterial cell envelope.61
CONCLUSIONS
We performed a quantitative census of NTA in pathogenic Mycobacterium to further understand the role of NTA in the physiology and pathogenicity of mycobacteria. We designed and validated a single-tube FASP-compatible enrichment of protein termini from complex microbial proteomes. We selectively enriched biologically N-terminally acetylated and chemically acetylated peptides from protein termini versus neo-peptides generated by protease digestion. The peptide populations were separated based on differences in their relative charge by strong-cation/anion exchange and further distinguished by the presence of light (H3) or heavy (D3) acetyl groups at the termini. In doing so, we defined 1336 protein termini across the M. tb and M. marinum proteomes; 42% of these polypeptides were acetylated at their N-terminus.
Overall, our findings indicate that NTA is widespread in mycobacterial pathogens. Our study identified the unique aspects of NTA in Mycobacterium, including the amino acid preference for NTA, and the fact that a subset of N-terminal proteins have differentially regulated levels of acetylation, some of which correlate with cellular localization. Together, these findings indicate that novel biology underlies NTA and its regulation, which may play widespread roles in modulating protein function in mycobacteria. The predominance of NTA of virulence-associated proteins suggests a new source of antimicrobial targets for drug discovery. Given the crucial role of acyl donors in physiology and virulence, targeting NTA pathways may provide additional avenues to combat mycobacterial disease.31,32
Supplementary Material
ACKNOWLEDGMENTS
The research findings reported in this study were supported by the National Institute of Allergies and Infectious Diseases of the National Institutes of Health under award number R01AI106872 to P.A.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Mass Spectrometry and Proteomics facility at the University of Notre Dame for their assistance.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00373.
Figure S-1. Arg frequency in canonical proteomes. Figure S-2. Comparison and analysis of M. tb H37Rv N-terminally acetylated proteins identified within the Kelkar et al. data set (2011). Figure S-3. Functional characterization of M. marinum N-terminally acetylated proteins. Figure S-4. IceLogo analysis of N-terminally acetylated protein populations for M. marinum. Figure S-5. IceLogo analysis of nonacetylated protein populations for M. tb. Figure S-6. IceLogo analysis of nonacetylated protein populations for M. marinum. Figure S-7. IceLogo analysis of noncanonical protein NTA for M. tb. Supplemental Methods. (PDF)
Supplemental Table 1. N-terminally acetylated proteins identified in M. tb Erdman. (XLSX)
Supplemental Table 2. N-terminally acetylated proteins identified in M. tb. (XLSX)
Supplemental Table 3. N-terminally acetylated proteins in M. marinum M. (XLSX)
Supplemental Table 4. Noncanonical acetylated proteins in M. tb. (XLSX)
Supplemental Table 5. N-terminal acetylation quantified in M. tb. (XLSX)
Supplemental Table 6. N-terminal acetylation quantified in M. marinum. (XLSX)
Supplemental Table 7. Comparisons of acetylation in M. tb and M. marinum. (PDF)
The authors declare no competing financial interest.
Processed and RAW data are available at MassIVE at accession number MSV000081168 (ftp://massive.ucsd.edu/MSV000081168).
REFERENCES
- (1).Driessen HP; de Jong WW; Tesser GI; Bloemendal H The mechanism of N-terminal acetylation of proteins. CRC Crit Rev. Biochem 1985, 18 (4), 281–325. [DOI] [PubMed] [Google Scholar]
- (2).Bradshaw RA; Brickey WW; Walker KW N-terminal processing: the methionine aminopeptidase and N alpha-acetyl transferase families. Trends Biochem. Sci 1998, 23 (7), 263–7. [DOI] [PubMed] [Google Scholar]
- (3).Starheim KK; Gevaert K; Arnesen T Protein N-terminal acetyltransferases: when the start matters. Trends Biochem. Sci 2012, 37 (4), 152–61. [DOI] [PubMed] [Google Scholar]
- (4). Polevoda B; Sherman F Nalpha-terminal acetylation of eukaryotic proteins. J. Biol. Chem 2000, 275 (47), 36479–82. [DOI] [PubMed] [Google Scholar]
- (5). Polevoda B; Sherman F N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol 2003, 325 (4), 595–622. [DOI] [PubMed] [Google Scholar]
- (6). Dikiy I; Eliezer D N-terminal acetylation stabilizes N-terminal helicity in lipid- and micelle-bound alpha-synuclein and increases its affinity for physiological membranes. J. Biol. Chem 2014, 289 (6), 3652–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7). Shemorry A; Hwang CS; Varshavsky A Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 2013, 50 (4), 540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8). Skoumpla K; Coulton AT; Lehman W; Geeves MA; Mulvihill DP Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 2007, 120 (9), 1635–45. [DOI] [PubMed] [Google Scholar]
- (9). Scott DC; Monda JK; Bennett EJ; Harper JW; Schulman BA N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 2011, 334 (6056), 674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10). Vetting MW; Bareich DC; Yu M; Blanchard JS Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008, 17 (10), 1781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11). Yoshikawa A; Isono S; Sheback A; Isono K Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol. Gen Genet 1987, 209 (3), 481–8. [DOI] [PubMed] [Google Scholar]
- (12). Arai K; Clark BF; Duffy L; Jones MD; Kaziro Y; Laursen RA; L’Italien J; Miller DL; Nagarkatti S; Nakamura S; Nielsen KM; Petersen TE; Takahashi K; Wade M Primary structure of elongation factor Tu from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 1980, 77 (3), 1326–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13). Tanaka S; Matsushita Y; Yoshikawa A; Isono K Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet 1989, 217 (2−3), 289–93. [DOI] [PubMed] [Google Scholar]
- (14). Itoh T Primary structure of an acidic ribosomal protein from Micrococcus lysodeikticus. FEBS Lett. 1981, 127 (1), 67–70. [DOI] [PubMed] [Google Scholar]
- (15). Bienvenut WV; Giglione C; Meinnel T Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics 2015, 15(14), 2503–18. [DOI] [PubMed] [Google Scholar]
- (16). Ouidir T; Jarnier F; Cosette P; Jouenne T; Hardouin J Characterization of N-terminal protein modifications in Pseudomonas aeruginosa PA14. J. Proteomics 2015, 114, 214–25. [DOI] [PubMed] [Google Scholar]
- (17). Kentache T; Jouenne T; De E; Hardouin J Proteomic characterization of Nalpha- and Nepsilon-acetylation in Acinetobacter baumannii. J. Proteomics 2016, 144, 148–58. [DOI] [PubMed] [Google Scholar]
- (18). Schmidt A; Kochanowski K; Vedelaar S; Ahrne E; Volkmer B; Callipo L; Knoops K; Bauer M; Aebersold R; Heinemann M The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol 2016, 34 (1), 104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19). Bonissone S; Gupta N; Romine M; Bradshaw RA; Pevzner PA N-terminal protein processing: A comparative proteogenomic analysis. Mol. Cell. Proteomics 2013, 12 (1), 14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20). Koch R Tuberculosis etiology. Dtsch. Gesundheitswes 1884, 7(15), 457–65. [PubMed] [Google Scholar]
- (21). WHO. WHO Global TB Report 2016, 2016. http://apps.who.int/medicinedocs/en/d/Js23098en/.
- (22). WHO. The Top Ten Causes of Death, 2017. http://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- (23). Nunes-Costa D; Alarico S; Dalcolmo MP; Correia-Neves M; Empadinhas N The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis (Oxford,U. K.) 2016, 96, 107–19. [DOI] [PubMed] [Google Scholar]
- (24). Tortoli E Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev 2014, 27(4), 727–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25). Tobin DM; Ramakrishnan L Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell. Micro-biol. 2008, 10 (5), 1027–39. [DOI] [PubMed] [Google Scholar]
- (26). Stinear TP; Seemann T; Harrison PF; Jenkin GA; Davies JK; Johnson PD; Abdellah Z; Arrowsmith C; Chillingworth T; Churcher C; Clarke K; Cronin A; Davis P; Goodhead I; Holroyd N; Jagels K; Lord A; Moule S; Mungall K; Norbertczak H; Quail MA; Rabbinowitsch E; Walker D; White B; Whitehead S; Small PL; Brosch R; Ramakrishnan L; Fischbach MA; Parkhill J; Cole ST Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 18 (5), 729–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27). Stamm LM; Brown EJ Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004, 6 (15), 1418–28. [DOI] [PubMed] [Google Scholar]
- (28). Shiloh MU; Champion PA To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr. Opin. Microbiol 2010, 13 (1), 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29). Volkman HE; Clay H; Beery D; Chang JC; Sherman DR; Ramakrishnan L Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004, 2 (11), e367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30). Kelkar DS; Kumar D; Kumar P; Balakrishnan L; Muthusamy B; Yadav AK; Shrivastava P; Marimuthu A; Anand S; Sundaram H; Kingsbury R; Harsha HC; Nair B; Prasad TS; Chauhan DS; Katoch K; Katoch VM; Kumar P; Chaerkady R; Ramachandran S; Dash D; Pandey A Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell. Proteomics 2011, 10 (12), M111.011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31). Vetting MW; Magnet S; Nieves E; Roderick SL; Blanchard JS A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol 2004, 11 (4), 565–73. [DOI] [PubMed] [Google Scholar]
- (32). Favrot L; Blanchard JS; Vergnolle O Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation. Biochemistry 2016, 55 (7), 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33). Rawat M; Av-Gay Y Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol Rev. 2007, 31 (3), 278–92. [DOI] [PubMed] [Google Scholar]
- (34). Mba Medie F; Champion MM; Williams EA; Champion PA Homeostasis of N-alpha terminal acetylation of EsxA correlates with virulence in Mycobacterium marinum. Infect. Immun 2014, 82(11), 4572–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35). Reyna C; Mba Medie F; Champion MM; Champion PA Rational engineering of a virulence gene from Mycobacterium tuberculosis facilitates proteomic analysis of a natural protein N-terminus. Sci. Rep 2016, 6, 33265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36). Okkels LM; Muller EC; Schmid M; Rosenkrands I; Kaufmann SH; Andersen P; Jungblut PR CFP10 discriminates between nonacetylated and acetylated ESAT-6 of Mycobacterium tuberculosis by differential interaction. Proteomics 2004, 4 (10), 2954–60. [DOI] [PubMed] [Google Scholar]
- (37). Lange S; Rosenkrands I; Stein R; Andersen P; Kaufmann SH; Jungblut PR Analysis of protein species differentiation among mycobacterial low-Mr-secreted proteins by narrow pH range Immobiline gel 2-DE-MALDI-MS. J. Proteomics 2014, 97, 235–44. [DOI] [PubMed] [Google Scholar]
- (38). Champion PA; Champion MM; Manzanillo P; Cox JS ESX-1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Mol. Microbiol 2009, 73(5), 950–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39). Champion MM; Williams EA; Kennedy GM; Champion PA Direct detection of bacterial protein secretion using whole colony proteomics. Mol. Cell. Proteomics 2012, 11, 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40). Champion MM; Williams EA; Pinapati RS; Champion PA Correlation of Phenotypic Profiles Using Targeted Proteomics Identifies Mycobacterial Esx-1 Substrates. J. Proteome Res. 2014, 13(11), 5151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41). Li Y; Champion MM; Sun L; Champion PA; Wojcik R; Dovichi NJ Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry as an Alternative Proteomics Platform to Ultraperformance Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry for Samples of Intermediate Complexity. Anal. Chem 2012, 84 (3), 1617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42). Stanley SA; Raghavan S; Hwang WW; Cox JS Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (22), 13001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43). Guinn KM; Hickey MJ; Mathur SK; Zakel KL; Grotzke JE; Lewinsohn DM; Smith S; Sherman DR Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol 2004, 51 (2), 359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44). Abdallah AM; Verboom T; Hannes F; Safi M; Strong M; Eisenberg D; Musters RJ; Vandenbroucke-Grauls CM; Appelmelk BJ; Luirink J; Bitter W A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol 2006, 62 (3), 667–79. [DOI] [PubMed] [Google Scholar]
- (45). Abdallah AM; Savage ND; van Zon M; Wilson L; Vandenbroucke-Grauls CM; van der Wel NN; Ottenhoff TH; Bitter W The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J. Immunol 2008, 181 (10), 7166–75. [DOI] [PubMed] [Google Scholar]
- (46). Bottai D; Majlessi L; Simeone R; Frigui W; Laurent C; Lenormand P; Chen J; Rosenkrands I; Huerre M; Leclerc C; Cole ST; Brosch R ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J. Infect. Dis 2011, 203 (8), 1155–64. [DOI] [PubMed] [Google Scholar]
- (47). Meinnel T; Giglione C Tools for analyzing and predicting N-terminal protein modifications. Proteomics 2008, 8 (4), 626–49. [DOI] [PubMed] [Google Scholar]
- (48). Lai ZW; Petrera A; Schilling O Protein amino-terminal modifications and proteomic approaches for N-terminal profiling. Curr. Opin. Chem. Biol 2015, 24, 71–9. [DOI] [PubMed] [Google Scholar]
- (49). Van Damme P; Arnesen T; Gevaert K Protein alpha-N-acetylation studied by N-terminomics. FEBS J. 2011, 278 (20), 3822–34. [DOI] [PubMed] [Google Scholar]
- (50). Hartmann EM; Armengaud J N-terminomics and proteogenomics, getting off to a good start. Proteomics 2014, 14 (23−24), 2637–46. [DOI] [PubMed] [Google Scholar]
- (51). Guryca V; Lamerz J; Ducret A; Cutler P Qualitative improvement and quantitative assessment of N-terminomics. Proteomics 2012, 12 (8), 1207–16. [DOI] [PubMed] [Google Scholar]
- (52). Marino G; Eckhard U; Overall CM Protein Termini and Their Modifications Revealed by Positional Proteomics. ACS Chem. Biol 2015, 10 (8), 1754–64. [DOI] [PubMed] [Google Scholar]
- (53). Zhao Y; Sun L; Champion MM; Knierman MD; Dovichi NJ Capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry for top-down characterization of the Mycobacterium marinum secretome. Anal. Chem 2014, 86 (10), 4873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54). Rogers LD; Overall CM Proteolytic post-translational modification of proteins: proteomic tools and methodology. Mol. Cell. Proteomics 2013, 12 (12), 3532–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55). Staes A; Van Damme P; Helsens K; Demol H; Vandekerckhove J; Gevaert K Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 2008, 8 (7), 1362–70. [DOI] [PubMed] [Google Scholar]
- (56). Gevaert K; Goethals M; Martens L; Van Damme J; Staes A; Thomas GR; Vandekerckhove J Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol 2003, 21 (5), 566–9. [DOI] [PubMed] [Google Scholar]
- (57). Staes A; Impens F; Van Damme P; Ruttens B; Goethals M; Demol H; Timmerman E; Vandekerckhove J; Gevaert K Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc 2011, 6 (8), 1130–41. [DOI] [PubMed] [Google Scholar]
- (58). Venne AS; Solari FA; Faden F; Paretti T; Dissmeyer N; Zahedi RP An improved workflow for quantitative N-terminal charge-based fractional diagonal chromatography (ChaFRADIC) to study proteolytic events in Arabidopsis thaliana. Proteomics 2015, 15(14), 2458–69. [DOI] [PubMed] [Google Scholar]
- (59). Lai ZW; Gomez-Auli A; Keller EJ; Mayer B; Biniossek ML; Schilling O Enrichment of protein N-termini by charge reversal of internal peptides. Proteomics 2015, 15 (14), 2470–8. [DOI] [PubMed] [Google Scholar]
- (60). Chen L; Shan Y; Weng Y; Sui Z; Zhang X; Liang Z; Zhang L; Zhang Y Hydrophobic Tagging-Assisted N-Termini Enrichment for In-Depth N-Terminome Analysis. Anal. Chem 2016, 88 (17), 8390–5. [DOI] [PubMed] [Google Scholar]
- (61). Williams EA; Mba Medie F; Bosserman RE; Johnson BK; Reyna C; Ferrell MJ; Champion MM; Abramovitch RB; Champion PA A Nonsense Mutation in Mycobacterium marinum That Is Suppressible by a Novel Mechanism. Infect. Immun 2017, 85(2), e00653–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62). Champion PA; Stanley SA; Champion MM; Brown EJ; Cox JS C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 2006, 313 (5793), 1632–6. [DOI] [PubMed] [Google Scholar]
- (63). James GT Inactivation of the protease inhibitor phenylmethylsulfonyl fluoride in buffers. Anal. Biochem 1978, 86 (2), 574–9. [DOI] [PubMed] [Google Scholar]
- (64). Shilov IV; Seymour SL; Patel AA; Loboda A; Tang WH; Keating SP; Hunter CL; Nuwaysir LM; Schaeffer DA The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 2007, 6 (9), 1638–55. [DOI] [PubMed] [Google Scholar]
- (65). Kapopoulou A; Lew JM; Cole ST The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Oxford, U. K.) 2011, 91 (1), 8–13. [DOI] [PubMed] [Google Scholar]
- (66). Elias JE; Gygi SP Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4 (3), 207–14. [DOI] [PubMed] [Google Scholar]
- (67). Colaert N; Helsens K; Martens L; Vandekerckhove J; Gevaert K Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 2009, 6 (11), 786–7. [DOI] [PubMed] [Google Scholar]
- (68). Swaney DL; Wenger CD; Coon JJ Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 2010, 9 (3), 1323–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69). Tsiatsiani L; Heck AJ Proteomics beyond trypsin. FEBS J. 2015, 282 (14), 2612–26. [DOI] [PubMed] [Google Scholar]
- (70). Wisniewski JR; Zougman A; Nagaraj N; Mann M Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6 (5), 359–62. [DOI] [PubMed] [Google Scholar]
- (71). Karp PD; Ouzounis CA; Moore-Kochlacs C; Goldovsky L; Kaipa P; Ahren D; Tsoka S; Darzentas N; Kunin V; Lopez-Bigas N Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005, 33 (19), 6083–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72). Zapf J; Sen U; Madhusudan; Hoch JA; Varughese KI A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction. Structure 2000, 8 (8), 851–62. [DOI] [PubMed] [Google Scholar]
- (73). Majlessi L; Prados-Rosales R; Casadevall A; Brosch R Release of mycobacterial antigens. Immunol Rev. 2015, 264 (1), 25–45. [DOI] [PubMed] [Google Scholar]
- (74). Bosserman RE; Champion PA ESX systems and the Mycobacterial Cell Envelope: What’s the connection? J. Bacteriol 2017, 199 (17), e00131–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75). Houben EN; Korotkov KV; Bitter W Take five - Type VII secretion systems of Mycobacteria. Biochim. Biophys. Acta, Mol. Cell Res 2014, 1843 (8), 1707–16. [DOI] [PubMed] [Google Scholar]
- (76). Champion MM; Campbell CS; Siegele DA; Russell DH; Hu JC Proteome analysis of Escherichia coli K-12 by two-dimensional native-state chromatography and MALDI-MS. Mol. Microbiol 2003, 47 (2), 383–96. [DOI] [PubMed] [Google Scholar]
- (77). Pathak D; Bhat AH; Sapehia V; Rai J; Rao A Biochemical evidence for relaxed substrate specificity of Nalphaacetyltransferase (Rv3420c/rimI) of Mycobacterium tuberculosis. Sci. Rep 2016, 6, 28892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78). Varshavsky A The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U. S. A 1996, 93 (22), 12142–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79). Li GW; Burkhardt D; Gross C; Weissman JS Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 2014, 157 (3), 624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80). Chittum HS; Champney WS Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr. Microbiol 1995, 30 (5), 273–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.