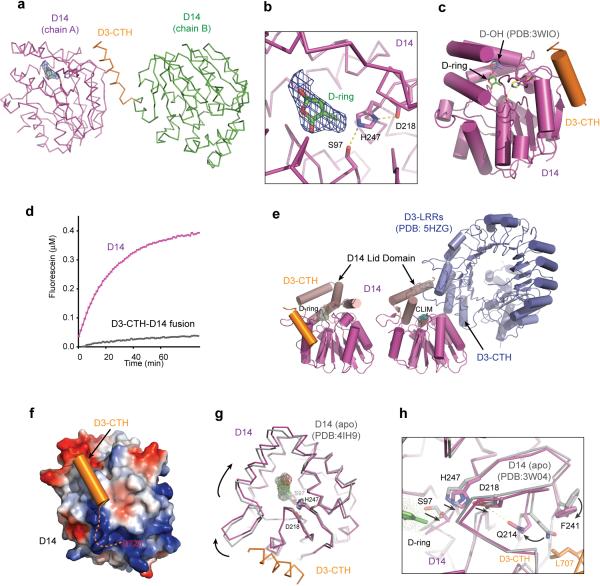
Extended Data Figure 5. Structural Analysis of D3-CTH-D14-GR24 complex.
(a) Packing of two D14 molecules N-terminally fused with D3-CTH. The D3-CTH region in Chain A is omitted. The GR24 D-ring (sticks) is shown together with the surround 2Fo-Fc electron density map calculated before the compound modeled in and contoured at 0.8σ. (b) A close-up view of the GR24 D-ring (sticks, green) and its electron density calculated as in (a). (c) Overall structure of D3-CTH (orange)-bound D14 (magenta) with a GR24 D-ring (sticks, green). The GR24 hydrolysis product, D-OH (sticks, cyan), revealed in the D14-D-OH structure (PDB: 3WIO) is shown based on superposition analysis. (d) Kinetics of YLG hydrolysis by free and D3-CTH-fused D14 (repeated thrice). (e) Comparison of the interface D14 (magenta and brown) makes upon binding to D3-CTH (orange) vs. ASK1-D3 (blue). The lid domain (brown) of D14 adopts open and closed conformation upon binding to D3-CTH and ASK1-D3, respectively. (f) Electrostatic potential surface map of D14 with D3-CTH (orange) bound. Dashed line indicates the otherwise free C-terminal region of D3 if D3-CTH were not fused to another copy of D14 in the crystal. (g) Conformational changes in the D14 lid domain induced by D3-CTH binding as revealed by superposition analysis between D3-CTH-bound (magenta) and apo D14 (grey, PDB: 4IH9). Arrows indicate the rotation of the D14 lid domain induced by D3-CTH (orange) relative to the catalytic triad shown in sticks. (h) Superposition analysis of D3-CTH-bound and apo D14 (PDB: 3W04) highlighting a possible allosteric pathway connecting Leu707 of D3-CTH to the catalytic triad of D14. Arrows indicate conformational changes within D14 induced by D3-CTH binding.