Fig. 1.
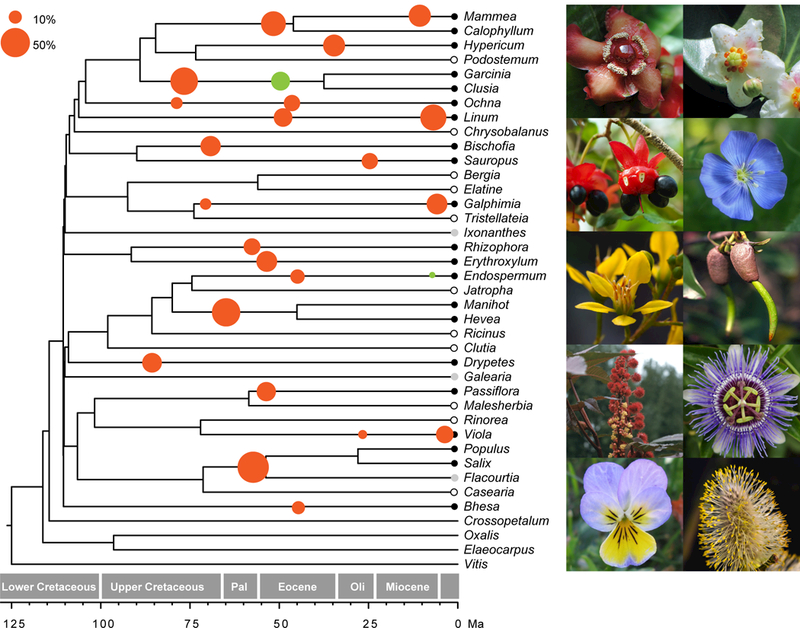
Phylogenetic distribution of whole genome duplications (WGDs) in Malpighiales. Species tree of Malpighiales inferred from 5113 gene trees using a summary coalescent method. WGDs identified from Ks analysis are illustrated with solid black dots on the terminal branches of corresponding species; species indicated with grey dots have transcriptomes that are potentially insufficient for adequate assessment of WGD using the Ks method. Two Salix species are collapsed into one terminal branch for simplicity. Circles illustrated along branches are dated WGDs (divergence time of parental genomes) from our phylogenomic reconciliation analysis. The radius of each circle is proportional to the percentage of orthologus genes supporting the WGD as determined from phylogenomic reconciliation (scale, top left). Solid red circles are significant WGDs as confirmed by our gene-count analysis; solid green circles indicate WGDs that do not receive significant support using the gene count method. Photos on the right are representatives of major Malpighiales clades sampled for investigating whole genome duplications, including from top to bottom (beginning with the left column): Garcinia, Ochna, Tristellateia, Manihot, Viola, Clusia, Linum, Rhizophora, Passiflora, and Salix. Ma, million years ago.
