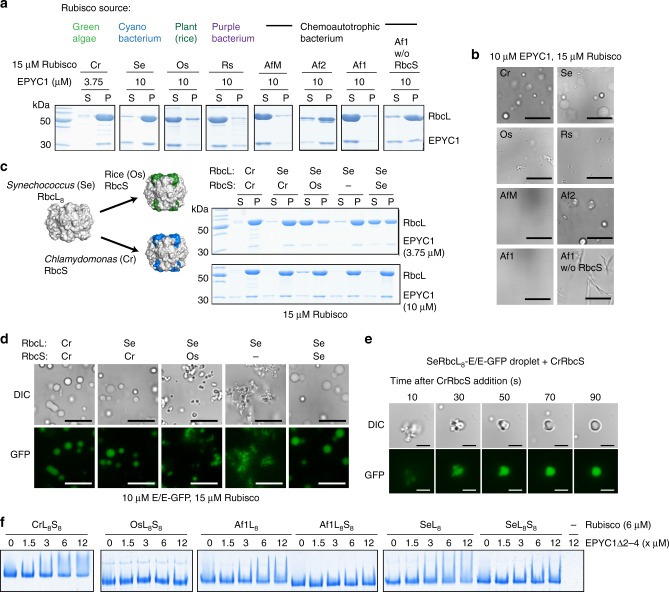
Fig. 4.
Diverse Rubisco enzymes vary in their tendency to demix with EPYC1. a, b The tendency of a phylogenetically diverse set of Rubisco enzymes to demix was assessed using the sedimentation assay (a) and microscopy (b). Scale bar 15 µm. c, d The role of the small subunit in demixing was assessed using chimeric Rubiscos (Synechococcus large subunit cores (L8) either lacking, or assembled with small subunits from rice (Os) or Chlamydomonas (Cr). Sedimentation (c) and microscopy (d) assays are shown. Scale bar 15 µm. e Addition of external CrRbcS (4 µM) rapidly changes the appearance of SeL8 (2.5 µM)-EPYC1/EPYC1-GFP (1.25/0.1 µM) droplets. Scale bar 5 µm. f Native PAGE gel-shift assay using Rubisco and the single-repeat variant EPYC1Δ2-4 indicates increased binding for Rubiscos lacking the small subunit