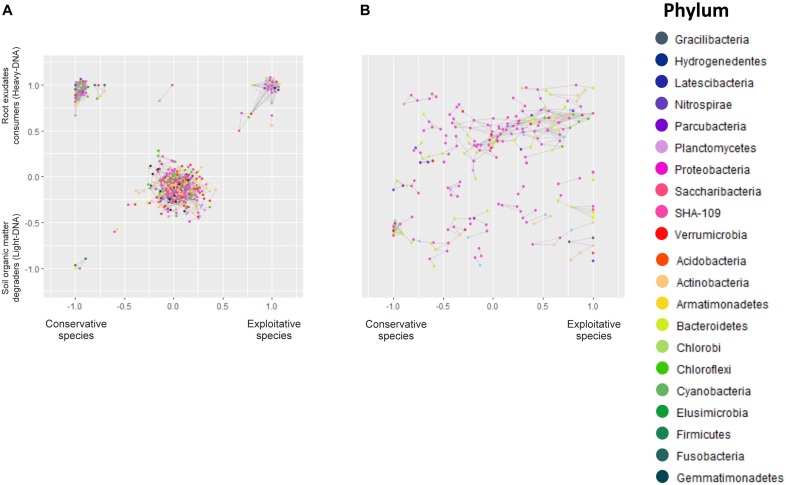
FIGURE 5.
Microbial networks of bacteria inhabiting (A) the RAS and involved in root exudate assimilation (heavy-DNA) or SOM degradation (light-DNA). Microbial networks of bacteria colonizing (B) the root tissues of conservative plants (Festuca paniculata and Sesleria caerulea) and exploitative plants (Bromus erectus, Anthoxanthum odoratum, Dactylis glomerata, and Trisetum flavescens). Each node represents a gene rRNA sequence, whereas the color indicates the phylum. Bacteria co-occurrence patterns, represented by gray lines, were assessed using a Bray–Curtis distance based on the repartition of OTUs in different plants and compartments. The resulting networks are positioned on a graph representing the gradient from conservative to exploitative plant species on the horizontal axis, and the gradient from SOM degraders (light-DNA) to root exudate consumers (heavy-DNA) on the vertical axis in graph (A). Overlapping nodes were scattered.