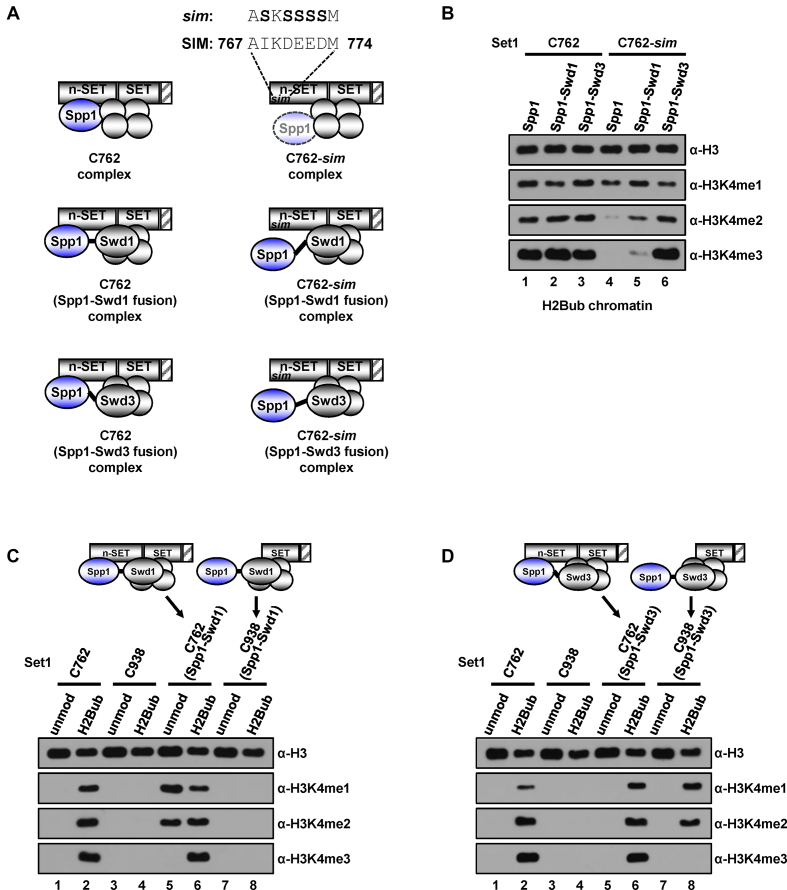
Figure 3.
H3K4 methylation activities of the subunit fusion-containing Set1 complexes. (A) Schematic representations of anticipated subunit associations in the purified complexes used in an in vitro HMT assay in (B). Four gray-colored circles indicate Swd1, Swd3, Bre2 and Sdc1. Amino acid substitutions to serine (bold) in the SIM (Spp1-interacting motif) are depicted in the C762-sim Set1 complex (upper right). In Spp1 and either Swd1 or Swd3 fusion proteins, two subunits are connected by the linker amino acids (GGGGGGAAA, depicted by short black lines). Depiction of faint Spp1 in the C762-sim Set1 complex implies its inefficient retention in the complex (Supplementary Figure S3A). (B) H3K4 methylation activities of Set1Cs containing physically-associated subunits. H2Bub chromatin templates were subjected to in vitro HMT assays with purified C762-based Set1 complexes containing subunit fusions (Supplementary Figure S3A). (C and D) Spp1-Swd1 fusion bypasses requirement of H2Bub for H3K4 methylation activity of the C762-based Set1 complex (C). Spp1-Swd3 fusion bypasses requirement of the n-SET domain for H2Bub-dependent H3K4 methylation activity of the C938-based Set1 complex (D). Chromatin templates assembled with unmodified H2B- or H2Bub-containing octamers were subjected to in vitro HMT assays with purified Set1Cs (Supplementary Figure S3B) that all have intact intrinsic H3K4 methyltransferase activities on free H3 (Supplementary Figure S3C). Subunit compositions and subunit fusions within the complexes are depicted at the top. unmod, unmodified H2B.