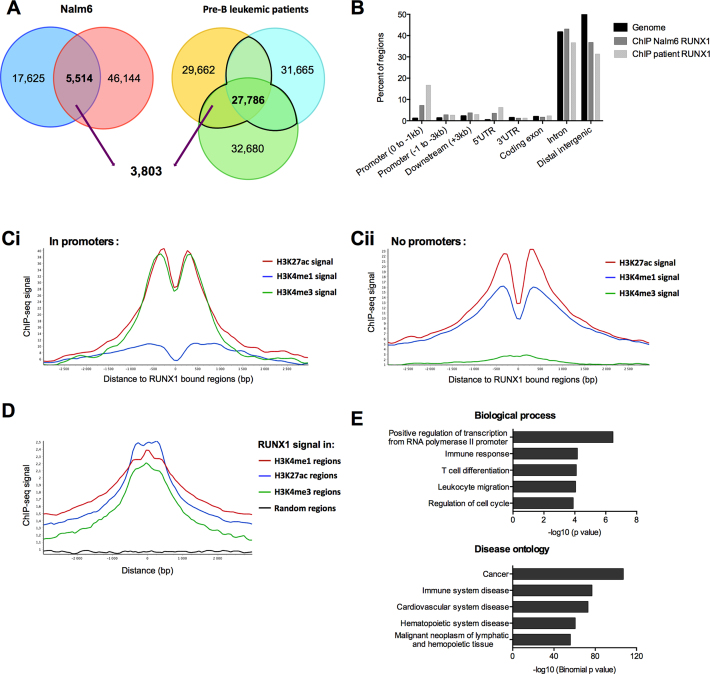
Figure 1.
RUNX1 ChIP-Seq signals are enriched within transcriptionally active chromatin regions. (A) Venn-diagrams presenting the intersection between RUNX1 binding regions in both replicates in Nalm6 cells, and bone marrow mononuclear cells isolated from three pre-B ALL patients. (B) Analyses of RUNX1 ChIP-Seq-enriched signals for pre-B Nalm6 cells and bone marrow mononuclear cells isolated from a pre-B acute lymphoblastic leukemia patient. ChIP-Seq reads were aligned to the reference human genome version GRCh37 (hg19). Analyses with the cis-regulatory Element Annotation System (CEAS) (47) software of RUNX1 ChIP-Seq provided RUNX1-bound regions enrichment in different genomic regions compared to the whole genome. (Ci and Cii) Read density plots for H3K4me1, H3K4me3 and H3K27ac enrichment centered around RUNX1 binding sites over a 3 kb window from RUNX1 Chip-Seq in Nalm6 cells, in promoter regions ([–4000 +1000] from TSS of annotated genes) (Ci) or without promoter regions (Cii). (D) Read density plots for RUNX1 signal around H3K4me1, H3K27ac, H3K4me3 regions over a 3 kb window and random regions of similar lengths in Nalm6 cells. The graph shows that the average RUNX1 signal peaked at regions positive for H3K4me3, H3K4me1 and H3K27ac. (E) Annotation enrichments of the 2651 potential RUNX1 target genes identified with DAVID (81) for biological process and GREAT (48) for disease ontology were shown. We selected 5 non-redundant annotations among the 10 most significant.