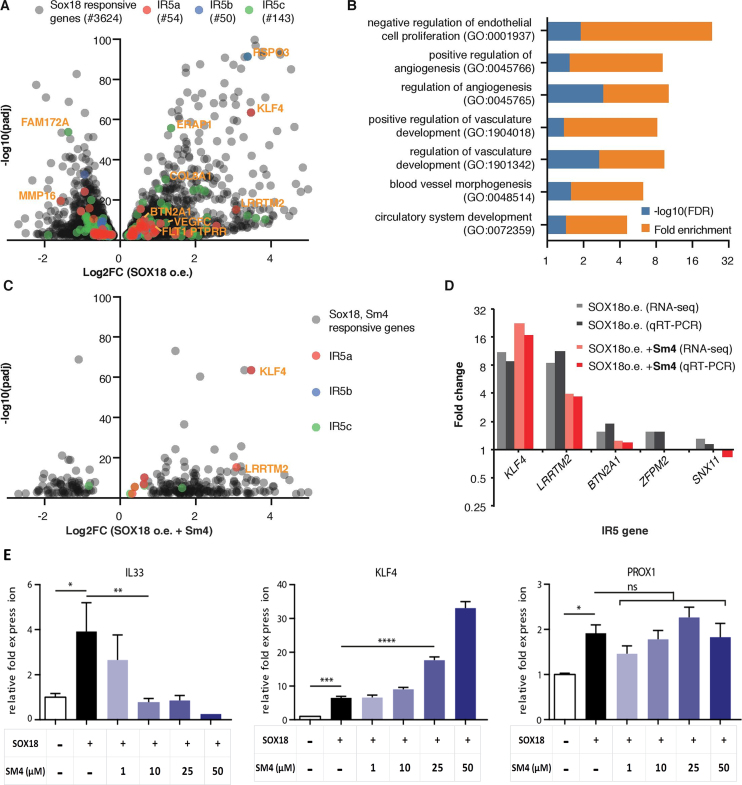
Figure 6.
Transcriptional endothelial signature of the SOX18 homodimer. (A) Volcano plot showing genes responsive to SOX18 over-expression (o.e.) in HUVECs. Genes with an IR5 motif have been highlighted in red (IR5a), blue (IR5b) and green (IR5c) and a selection of highly responsive genes has been annotated (e.g. KLF4). (B) GO term analysis (molecular function, PANTHER v13.1 (67)) filtered for vascular and endothelial terms for SOX18 responsive IR5 genes, revealing significant enrichment of up to 21.3 fold (Fisher test, FDR < 0.05). More comprehensive GO analysis for IR5 vs. non IR5 genes is presented in Supplementary Figure S10. (C) Volcano plot showing genes responsive to both SOX18 o.e. and to treatment with inhibitor Sm4. IR5 genes are labelled as in (A). Most Sm4 responsive IR5 genes are positively regulated by SOX18, including KLF4 and LRRTM2. (D) Validation of IR5 gene regulation using representative SOX18, Sm4 responsive IR5 genes. Data from qRT-PCR matches RNA-seq results and shows positive relation of IR5 genes by SOX18 o.e. and impact of SOX18 inhibition. (E) Dose response of Sm4 treatment (1–50 μM) by qRT-PCR analysis shows inhibition or over-expression of IL33 and KLF4 transcripts, respectively. PROX1 mRNA was not significantly affected. Error bars are s.e.m.