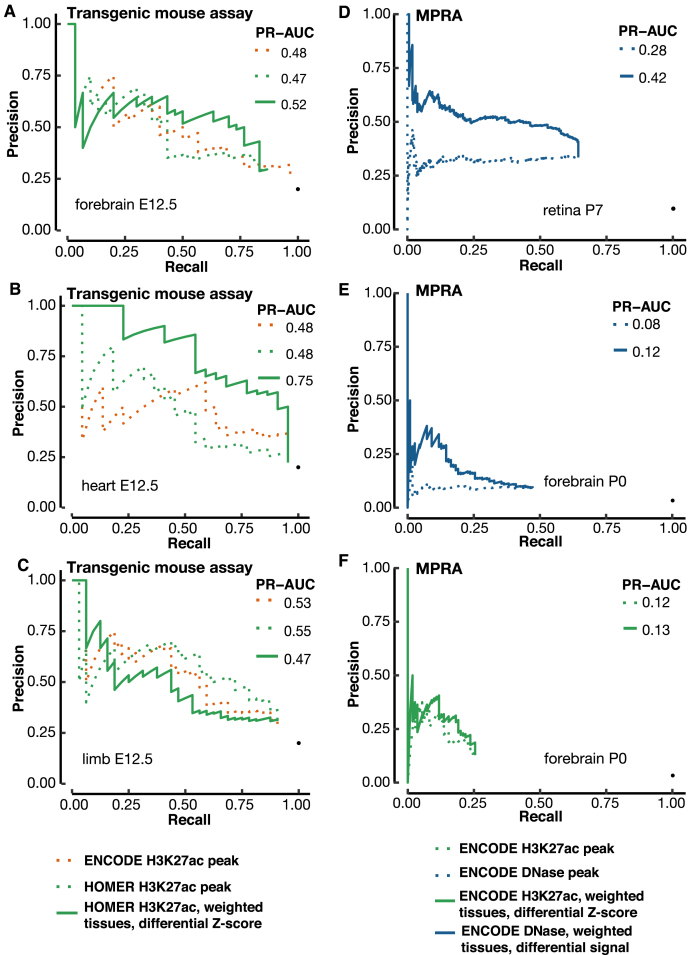
Figure 6.
Validation of our differential signal approaches with a blind test and MPRA enhancers. (A–C) A blind test of the differential H3K27ac signal method for predicting e12.5 transgenic mouse enhancers in (A) forebrain, (B) heart and (C) limb. We used 17 contrast tissues as in Figure 5, weighted proportionally to their distances from the tissue under investigation. VISTA enhancers in the tissue of interest were used for evaluating the precision and recall. The predictions were submitted to the ENCODE Portal before the release of experimental results, and hence were a blind test. PR-AUC values are indicated in the legend in each panel. (D, E) Using the differential DNase signal for predicting MPRA CRE-seq enhancers in retina (D) and (E) cerebral cortex. (F) Using the differential H3K27ac signal for predicting MPRA CRE-seq enhancers in cerebral cortex. We used the same set of contrast tissues as in Figure 5, weighted proportionally to their distances from the tissue under investigation.