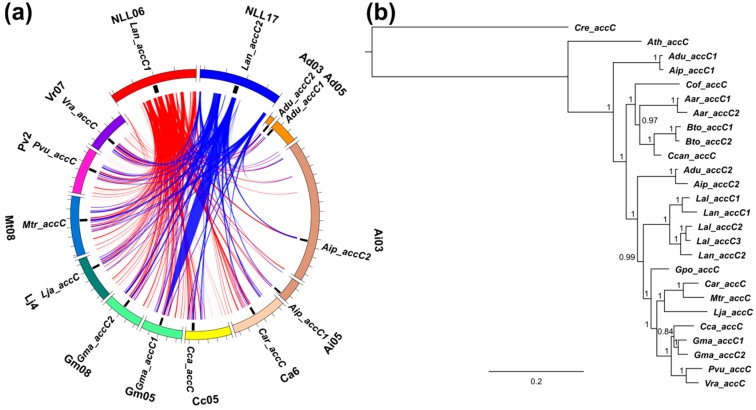
Figure 8.
Comparative mapping and phylogenetic inference of legume biotin carboxylase (accC) subunit genes of plastid ACCase. (a) syntenic patterns revealed for narrow-leafed lupin genome regions and corresponding regions of legume chromosomes. Abbreviated species names are provided as follows: Aar, Acacia argyrophylla; Adu, Arachis duranensis; Aip, A. ipaensis; Ath, Arabidopsis thaliana; Bto, Bauhinia tomentosa; Car, Cicer arietinum; Cca, Cajanus cajan; Ccan, Cercis canadensis; Cof, Copaifera officinalis; Cre, Chlamydomonas reinhardtii; Gma, Glycine max; Gpo, Gompholobium polymorphum; Lal, Lupinus albus; Lan, L. angustifolius; Lja, Lotus japonicus; Mtr, Medicago truncatula; Pvu, Phaseolus vulgaris; Vra, Vigna radiata. ACCase gene loci are indicated by black bars. Ribbons visualize homologous links identified by DNA sequence alignments. L. angustifolius genomic sequences and homology links are marked by colors: red and blue. (b) majority rule consensus trees found in a Bayesian analysis of legume accC genes. Numbers are posterior probabilities. The length of MAFFT alignment used for phylogenetic inference was 1716 nt.