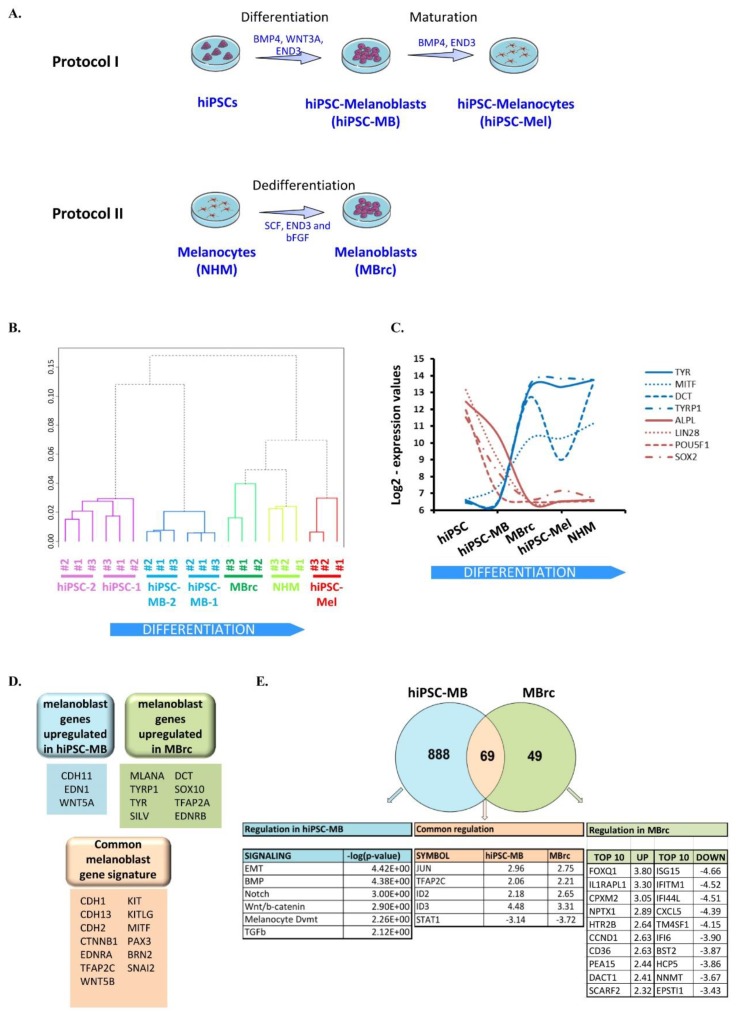
Figure 1.
Two different approaches to generate human melanoblasts. (A) Schematic of two protocols to generate human melanoblasts. Protocol I leads to the directed differentiation of human induced pluripotent stem cells (hiPSCs). Protocol II leads to the dedifferentiation of human primary melanocytes (NHM). (B) Dendrogram of hierarchical clustering of the transcriptome of the following samples: normal human melanocytes (NHM), human induced pluripotent stem cells (hiPSC-1 and hiPSC-2), hiPSC-derived melanocytes (hiPSC-Mel), hiPSC-derived melanoblasts (hiPSC-MB-1 and hiPSC-MB-2), and melanocyte-derived melanoblasts (MBrc). The function hcluster was used, in which the parameter correlation evokes computation of Pearson-type distances. (C) Log2-expression values of pluripotency genes (red curves) and melanocyte-specific genes (blue curves) in the same samples as in (B). Biological replicates were averaged. (D). Identification of known melanoblast markers, which are commonly or specifically expressed in each melanoblast population. (E) Gene set enrichment analysis with Ingenuity (IPA) of the transcriptome of both melanoblast populations, compared to NHM samples. Log2-threshold = 2, p-value < 0.05. The three lists of regulated genes are provided in Table S1.