Abstract
Background and Aims
An understanding of mycorrhizal variation, orchid seed germination temperature and the effect of co-occurring plant species could be critical for optimizing conservation translocations of endangered plants with specialized mycorrhizal associations.
Methods
Focusing on the orchid Thelymitra epipactoides, we isolated mycorrhizal fungi from ten plants within each of three sites; Shallow Sands Woodland (SSW), Damp Heathland (DH) and Coastal Heathland Scrub (CHS). Twenty-seven fungal isolates were tested for symbiotic germination under three 24 h temperature cycles: 12 °C for 16 h–16 °C for 8 h, 16 °C for 16 h–24 °C for 8 h or 27 °C constant. Fungi were sequenced using the internal transcribed spacer (ITS), nuclear large subunit 1 (nLSU1), nLSU2 and mitochondrial large rRNA gene (mtLSU). Orchids were grown to maturity and co-planted with each of ten associated plant species in a glasshouse experiment with tuber width measured at 12 months after co-planting.
Key Results
Two Tulasnella fungal lineages were isolated and identified by phylogenetic analyses, operational taxonomic unit 1 (OTU1) and ‘T. asymmetrica’. Fungal lineages were specific to sites and did not co-occur. OTU1 (from the SSW site) germinated seed predominantly at 12–16 °C (typical of autumn–winter temperature) whereas ‘T. asymmetrica’ (from the DH and CHS sites) germinated seed across all three temperature ranges. There was no difference in the growth of adult orchids germinated with different OTUs. There was a significant reduction in tuber size of T. epipactoides when co-planted with six of the commonly co-occurring plant species.
Conclusions
We found that orchid fungal lineages and their germination temperature can change with habitat, and established that translocation sites can be optimized with knowledge of co-occurring plant interactions. For conservation translocations, particularly under a changing climate, we recommend that plants should be grown with mycorrhizal fungi tailored to the recipient site.
Keywords: Orchidaceae, mycorrhizae, Tulasnella, translocation, Thelymitra epipactoides
INTRODUCTION
Globally, one in five plant species are under threat of extinction (Pimm et al., 2014; Anderson et al., 2016) from a diversity of threats such as habitat destruction (Tilman et al., 1994), invasive species (Mack et al., 2000), overexploitation (Brook et al., 2008) and climate change (Thomas et al., 2004). Human intervention is required to prevent extinctions, and increasingly conservation translocations (see definition in IUCNS, 2013, i.e. the deliberate movement of organisms intended to yield a conservation benefit at a population, species or ecosystem level) offer the last hope for species’ survival. One possible factor increasing the likelihood of extinction is specialization with a partner organism (Harcourt et al., 2002). Where specialists lose an essential symbiosis from part of their range, such as pollinators (Robertson et al., 1999; Anderson et al., 2011; Pauw and Hawkins, 2011; Reiter et al., 2017) or mycorrhizal fungi (Shefferson et al., 2005; Swarts et al., 2010), a range retraction is inevitable. Conversely, generalists are more likely to be resilient to losing a symbiotic partner as they may be able to switch to another association in part of their range (Johnson and Steiner, 1997). Therefore, conservation translocations of plants with specialized ecological interactions require not only a thorough knowledge of the threats and threat management (Vallee, 2004), but also a detailed understanding of the ecology (Reiter et al., 2016) of the pollinators (Phillips et al., 2014; Reiter et al., 2017) and mycorrhizal fungal associations (Batty et al., 2001). Yet specialized ecological interactions are rarely specifically addressed in plant conservation translocations (Reiter et al., 2016). Addressing this gap in both knowledge and conservation practice has the potential to increase the long-term success of plant translocations.
The Orchidaceae is a charismatic and diverse plant family with approx. 26 567 species globally (World Checklist of Selected Plant Families, 2017). With high levels of geographic endemism and complex relationships with other organisms (Swarts and Dixon, 2009; Orejuela-Gartner, 2012), it has been predicted that many orchids may be among the first to decline when a habitat is degraded (Backhouse, 2007). All orchids are reliant on mycorrhizal associations in order to germinate in the wild (Warcup, 1971; Rasmussen, 1995, 2002). However, their level of dependence may change throughout their life cycle (Rasmussen, 2002; Cameron et al., 2006). For example, there is evidence in some species that dependence on mycorrhizal fungi for nutrition and survival differs between seedlings and mature plants (Waterman and Bidartondo, 2008), achlorophyllous (Selosse and Roy, 2009) and chlorophyllous species (Stöckel et al., 2014). Furthermore, orchids differ in their specificity for mycorrhizal symbionts (Swarts et al., 2010), which may affect their predisposition to extinction, through limiting suitable sites at local or regional scales.
Orchids most commonly form mycorrhizal associations with fungi belonging to three groups (Tulasnellaceae, Sebacinaceae and Ceratobasidiaceae) of the basidiomycetes (Taylor et al., 2002; Weiss et al., 2004). The Tulasnellaceae are widespread orchid mycorrhizal fungi (OMFs) in both temperate and tropical regions (Dearnaley et al., 2012), and yet their ecology remains largely unstudied (Selosse and Martos, 2014); but see Fochi et al. (2017). Orchids forming mycorrhizal relationships with Tulasnellaceae may form relationships with either a phylogenetically narrow group of fungi, e.g. Cypripedium spp. (Shefferson et al., 2005, 2007) or a phylogenetically broad group of fungi (e.g. Tipularia discolor associations; McCormick et al., 2004).
Given their reliance on fungi for germination and in many cases annual growth, the distribution of OMFs could theoretically affect the geographic ranges of orchids. Orchids that are generalists (associating with a phylogenetically broad group of fungi), e.g. Dactylorhiza majalis (Jacquemyn et al., 2012), may have a broad distribution due to less specific niche requirements of their fungi. On the other hand, orchids that are specialists may have fewer fungal associations, but if the OMFs have a wide geographic distribution, e.g. in Pheladenia deformis (Davis et al., 2015), the orchid’s distribution is not limited by the OMF with which it associates. Those orchids that have a specific mycorrhizal relationship, but only associate with fungi that also have limited distribution, are likely to be restricted in range (Swarts et al., 2010), though in practice there has been limited evidence for orchid mycorrhiza with restricted ranges. Therefore, high mycorrhizal specificity (i.e. an orchid only forming symbiosis with one or a few lineages of mycorrhizal fungi from a narrow phylogenetic lineage) is not necessarily associated with orchid rarity. For example, narrow phylogenetic groups of tulasnelloid mycorrhizal fungi were found in threatened and common species of Drakaea (Phillips et al., 2011, 2014; Linde et al., 2014) and Chiloglottis (Roche et al., 2010; Linde et al., 2017), but because the OMFs are widely distributed (covering the range of the entire orchid genus), no impact on species distribution is expected. It is likely that factors other than OMF specificity and distribution impact on orchid distribution, as some rare orchids such as the highly endemic terrestrial Teagueia morphospecies in Ecuador, associate with a Tulasnellaceae OMF that has a worldwide distribution (Suárez et al., 2016).
To be confident that fungi associated with orchids are mycorrhizal, it is important to show an effective symbiosis between orchid and fungus. Although direct sequencing from orchid roots and soil allows for a complete picture of those organisms present, it does not enlighten us on the ability of these organisms to form an effective mycorrhizal association. Some caution is needed in the interpretation of patterns of specificity in previous papers, where the fungi identified have not been tested directly for the ability to form symbioses (see papers cited in the review of McCormick and Jacquemyn, 2014). More useful for orchid conservation are studies that both sequence and test germination ability (McCormick et al., 2004; Roche et al., 2010; Smith et al., 2010; Phillips et al., 2011; Linde et al., 2014; Mujica et al., 2016). In most orchids, we do not know if the mycorrhizal fungi that germinate seed also sustain adult plants. Tests for mycorrhizal compatibility with orchids typically are conducted with seed germination studies, where the production of rhizoids or a green leaf constitutes a positive interaction (Batty et al., 2001; Roche et al., 2010; Davis et al., 2015; Phillips et al., 2016). Longer term trials that compare the efficacy of different fungal species on orchid growth with maturation are rare, but showed that OMFs which induced the most seed to germinate were not necessarily those that resulted in the best survival of orchid seedlings (Rasmussen, 1995; Huynh et al., 2009). Furthermore, Smith et al. (2010) cautioned that long-term success in orchid survival under field conditions may be dependent on highly specific host–fungus interactions that may prove to be significant in the long term.
To date, the surrounding non-orchid flora’s effect on orchid growth, either detrimentally by competing for resources or positively by providing nutrients or suitable microclimate, is underexplored. If we are to conduct conservation translocations on species that are reliant upon mycorrhizal fungi for germination and to sustain populations in the wild, we require the fungi to be cultivable, able to germinate the seed and sustain adult plants. We need to understand whether these symbiotic associations change with different habitats and whether the ability of the mycorrhizal fungi to germinate orchid seed changes under different climatic conditions. Mycorrhizal distribution may be limited by edaphic conditions, including: soil chemistry, moisture, pH, organic content and nutrient availability (Perkins and McGee, 1995; McCormick et al., 2004; Bunch et al., 2013; McCormick and Jacquemyn, 2014). In arbuscular mycorrhiza, the associated vegetation or even invasive plant species may affect the mycorrhizal community abundance (Zubek et al., 2016). Therefore, tests of mycorrhizal symbiosis may benefit not only from the evaluation of short-term seed germination but also from longer term growth alone and with associated species from the sites where the orchids occur. We are not aware of any study with orchids that have utilized pot trials with naturally co-occurring plant species to investigate how they affect orchid growth.
Here we investigate aspects of mycorrhizal ecology useful for optimizing germination and conservation translocations. We use Thelymitra epipactoides (Orchidaceae), a species previously widespread across south-eastern mainland Australia, as our study species. Due to habitat destruction, T. epipactoides is now endangered under the federal Environment Protection and Biodiversity Conservation Act (1999) and is considered a high priority for conservation translocation (Coates et al., 2002). It now has a discontinuous distribution ranging from coastal to near-desert habitats across its range (Jeanes and Backhouse, 2006). Thelymitra epipactoides associates with Tulasnella asymmetrica based on morphological culture identification (Warcup, 1981). In order to optimize germination and conservation translocations, we investigate the OMF associations from T. epipactoides growing in ecologically and climatically different sites. Furthermore, we investigate whether OMFs from the different sites differ in their optimum temperature requirements for seed germination. Specifically, we ask the following questions. (1) Is there habitat-driven variation in Tulasnella associated with T. epipactoides? (2) Do Tulasnella operational taxonomic units (OTUs) from climatically different sites have different temperature requirements for orchid seed germination? (3) Do symbiotically grown T. epipactoides plants benefit from co-planting with associated flora?
MATERIALS AND METHODS
Study species
The genus Thelymitra consists of approx. 110 species, with a distribution across Australia, New Zealand, Indonesia, New Caledonia, New Guinea and the Philippines (Jeanes, 2013). Thelymitra epipactoides is a summer-dormant herbaceous geophyte that survives over summer as an underground tuber. Each tuber produces a single leaf and a flowering stalk to 18–30 cm tall with a raceme of between six and 25 flowers (Fig. 1) with a metallic lustre coloured pink, bronze, green, blue or red (Jeanes, 2011). The orchid is thought to be food deceptive and pollinated by native bees (Nomia and Lasioglossum sp.) (Cropper and Calder, 1990).
Fig. 1.
Mature plants of Thelymitra epipactoides grown during this study.
Sites
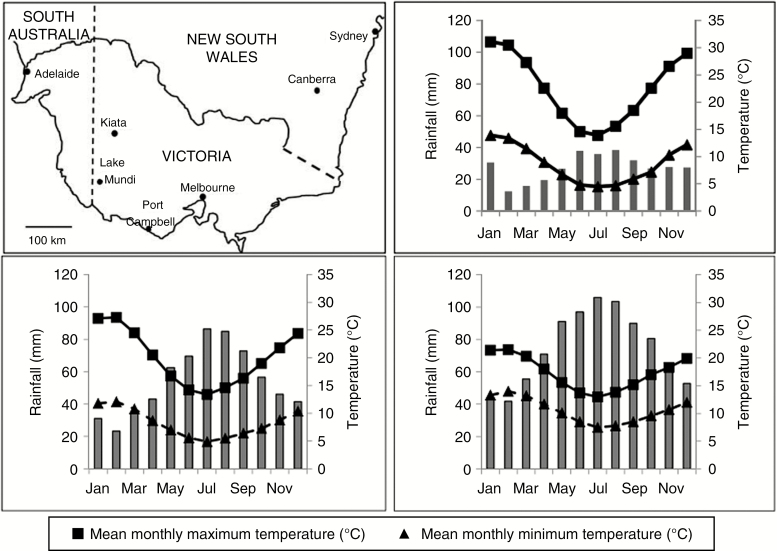
Thelymitra epipactoides populations growing in three habitats were studied: Shallow Sands Woodland (SSW), Damp Heathland (DH) and Coastal Heathland Scrub (CHS) (Table 1; Fig. 2). All three sites have a Mediterranean climate with cooler wetter winters and hotter drier summers. These sites represent the range of habitats and climate in which the species is known to occur, with variation in soil types, rainfall and temperature between sites. The SSW site is the driest with the most extreme temperature ranges, whereas the CHS site is the wettest with the least temperature range (Table 1; Fig. 2). The SSW vegetation is open with an overstorey of Eucalyptus leucoxylon, Callitris gracilis and Allocasuarina verticillata, a middle storey of a variety of acacias and a diverse understorey of Astroloma, bulbs including Arthropodium fimbriatum and Bulbine bulbosa, and 15 orchid species. The DH site is dense and consists predominantly of Leptospermum continentale and a variety of acacias with a diverse understorey of Lepidosperma, Stackhousia, Thomasia petalocalyx, Kennedia prostrata and a variety of legumes (Fabaceae). The CHS site is dense and consists of an overstorey of Melaleuca lanceolata and Allocasuarina verticillata, with a middle storey of Leptospermum continentale. The understorey included Poa poiformis, Lomandra longifolia, Correa reflexa and Dianella brevicaulis. The grass Rytidosperma occurs at all sites.
Table 1.
Vegetation type, location, number of Thelymitra epipactoides plants sampled across populations, Bureau of Meteorology weather station number (Australian Government Bureau of Meteorology, 2017), soil type, average annual rainfall, maximum daytime temperature, average temperature and minimum temperature of study sites
| Vegetation type | Location | No. of plants analysed | Weather station no. | Soil | Average annual rainfall (mm) | Maximum daytime temperature | Average daytime temperature | Minimum temperature |
|---|---|---|---|---|---|---|---|---|
| Shallow Sands Woodland | Kiata Flora and Fauna Reserve | 10 | 78015 | Deep sand | 327.1 | 46 | 22.6 | –5 |
| Damp Heathland | Lake Mundi Flora and Fauna Reserve | 10 | 90182 | Sandy loam | 695.2 | 44.5 | 20.1 | –3.1 |
| Coastal Heathland Scrub | Port Campbell National Park | 10 | 90015 | Clay loam | 905.1 | 43.3 | 17.3 | –1.1 |
Fig. 2.
Rainfall and temperature for each site from the closest weather stations of the Australian Government Bureau of Meteorology: Nhill for the SSW site (top right), Casterton for DHS (bottom left) and Cape Otway lighthouse for the CH site (bottom right). Bars = mean monthly rainfall (mm).
Fungal isolation
In winter, when plants were actively growing, ten plants from each site had sections of lateral roots above the tubers removed, and fungi were isolated from these roots within 4 h of collection. Roots were gently rubbed by hand and rinsed for 15 min in running water before selected regions were surface-sterilized in 0.05 % NaOCl for 3 min and rinsed with sterile water. Root tissue was sliced open with a glass blade to release pelotons. Pelotons were rinsed in a series of sterile water droplets (Rasmussen et al., 1990) in a laminar flow cabinet. Individual pelotons were plated onto fungal isolation medium (FIM) (Clements et al., 1986) containing 0.05 g L–1 streptomycin. Three isolates per plant were grown, of which 32 isolates were sequenced across all regions.
Fungal sequencing
Fungal isolates were sub-cultured into 40 mL aliquots of liquid oatmeal broth (25 g of rolled oats, boiled, strained and made up to 1 L with water) and grown in shake-culture at 25 °C for 4 weeks. Fungal pellets were removed from the liquid medium, blotted on tissue and DNA was extracted from approx. 100 mg of each fungus using a Qiagen Plant DNeasy kit according to the manufacturer’s instructions, including the optional centrifugation step (Qiagen, Hilden, Germany), and elution into 2 × 50 μL instead of 2 × 100 μL.
Sequences of four regions [internal transcribed spacer (ITS), nuclear large subunit 1 (nLSU1), nLSU2 and mitochondrial large rRNA gene (mtLSU)] were obtained using the primers and thermocycling conditions listed in Table 2. Thirty-two isolates across all sites were sequenced across all regions: with 13 isolates from SSH, eight from DH and 11 from CHS sites. For the initial PCR amplifications, each 25 μL reaction contained 12.5 μL of Promega GoTaq Green Master Mix, 8.5 μL of nuclease-free water, 1 μL each of forward and reverse primers (10–25 μm) and 2 μL containing 5–20 ng of genomic DNA or sterile nuclease-free water. After amplification in a G-Storm thermocycler using the PCR conditions in Table 2, a 5 μL aliquot of the products was electrophoresed at 80–100 V alongside a GeneRuler™ 100 bp Plus ladder in a 1.4 % agarose gel either in TBE [Tris-borate-EDTA: 10.8 g L–1 Tris base, 5.5 g L–1 boric acid, 4 mL of 0.5 m EDTA pH 8.0] or in TAE (Tris-acetate-EDTA: 4.84 g L–1 Tris base, 1.142 g L–1 glacial acetic acid, 2 mL of 0.5 m EDTA pH 8.0), stained with ethidium bromide and imaged using a Bio-Rad Gel Doc system with Quantity One software.
Table 2.
Primers and cycling conditions used for fungal DNA analysis
| PCR region/ type | Primers (5’–3’) | Initial denaturation | No. of cycles | Cycling parameters | Final extension | References | ||
|---|---|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||||
| ITS | ITS1: (TCCGTAGGTGAACCTGCGG) ITS4: (TCCTCCGCTTATTGATATGC) |
94 °C (10 min) | 35 | 94 °C (30 s) | 51 °C (30 s) | 72°C (1 min) | 72 °C (10 min) | White et al. (1990) |
| nLSU1 | Ctb6: (GCATATCAATAAGCGGAGG) TW13: (GGTCCGTGTTTCAAGACG) |
94 °C (10 min) | 35 | 94 °C (1 min) | 49 °C (1 min) | 72 °C (2 min) | 72 °C (10 min) | Bruns Lab (http://nature.berkeley.edu/ brunslab/tour/primers.htm) |
| nLSU2 | CTW13: (CGTCTTGAAACACGGACC) TW14: (GCTATCCTGAGGGAAACTTC) |
94 °C (10 min) | 35 | 94 °C (1 min) | 50 °C (1 min) | 72 °C (2 min) | 72 °C (10 min) | Bruns Lab (http://nature.berkeley.edu/ brunslab/tour/primers.htm) |
| MtLSU | ML5: (CTCGGCAAATTATCCTCATAAG) ML6: (CAGTAGAAGCTGCATAGGGTC) |
94 °C (10 min) | 35 | 94 °C (30 s) | 51 °C (30 s) | 72 °C (1 min) | 72 °C (10 min) | White et al. (1990) |
| Sequencing | ITS4/ML6/CTB6/TW13/CTW13/TW14 | – | 25 | 96 °C (10 s) | 50 °C (5 s) | 60 °C (4 min) | – | Applied Biosystems (www. thermofisher.com) |
ITS, nuclear ribosomal internal transcribed spacer region; nLSU1, nuclear large subunit (28S) region 1; nLSU2, nuclear large subunit (28S) region 2; and MtLSU, mitochondrial large RNA gene
The PCR products were purified using a Qiagen QIAquick PCR Purification Kit according to the manufacturer’s instructions. Purified products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, www.thermofisher.com). Each 20 μL reaction contained: 2 μL of 10× buffer, 14 μL of nuclease-free water, 1 μL of Big Dye mix (Version 3.1), 1 μL of the appropriate primer (0.16 μm) and 2 μL of cleaned PCR product (5–20 ng). Cycling parameters were as in Table 2. DNA was precipitated using the Applied Biosystems ethanol-precipitation protocol 1, and products were electrophoresed and sequenced at Micromon, Monash University (https://platforms.monash.edu/micromon/).
Analysis of DNA sequence data
Alignments of the four DNA sequence regions were performed in Geneious V10.0.8 (Kearse et al., 2012). A Maximum Likelihood analysis for each individual DNA region, as well as a concatenated four-region alignment, was run in RAxML (Stamatakis et al., 2008) through Geneious with 1000 bootstrap replicates. Furthermore, a Bayesian analysis was run in MrBayes 3.2.2 (Ronquist et al., 2012), also through Geneious. Analyses were run for 2 million generations (first 25 % discarded as burn-in, Markov chain sampled every 1000 generations) with four chains, using the GTR + G model. Although trees were rooted to T. albida, trees were later visualized and midpoint rooted in Figtree v 1.4 (Rambaut and Drummond, 2012). Convergence was considered to be sufficient if the s.d. of the split frequencies was <0.02.
The maximum level of sequence divergence at the ITS was calculated within a putative species using Geneious V10.0.8 (Kearse et al., 2012). To establish the level of divergence within species, minimum and maximum sequence divergences within a clade were calculated. Also, the maximum sequence divergence within a species was compared with the minimum genetic divergence with any of the individuals in the sister clade. Closely related sequences from GenBank were included in Figs 3 and 4.
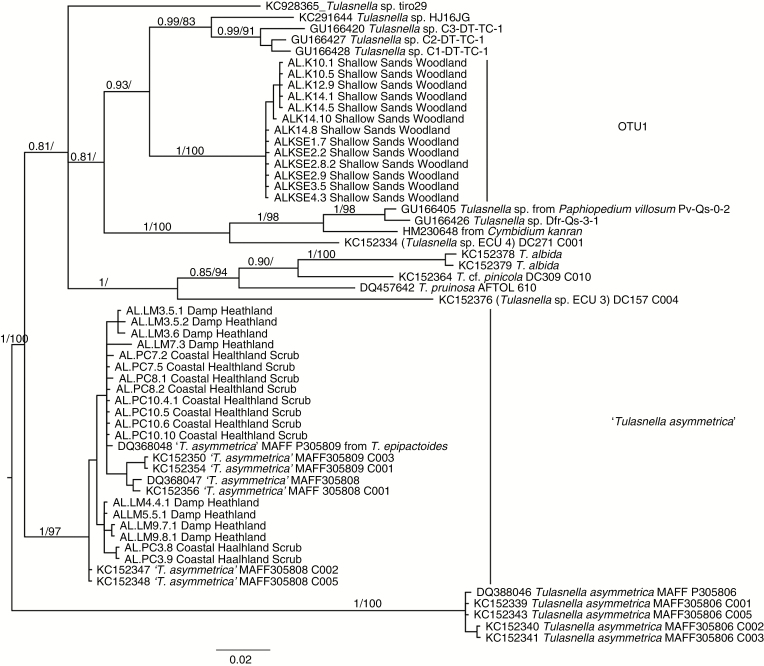
Fig. 3.
Phylogenetic tree for the ITS region of Tulasnella isolates from the Shallow Sands Woodland, Damp Sands Heathland and Coastal Heathland sites. The sequences clustered into two well-supported (bootstrap = 100 %) OTUs (‘Tulasnella asymmetrica’ and OTU1), based on a 5 % sequence divergence cut-off (Linde et al., 2014, 2017). Warcup’s (1981) isolates of T. asymmetrica were separated into T. asymmetrica and ‘T. asymmetrica’ based on phylogenetic divergence (Cruz et al., 2014). We have kept this separation and refer to ‘T. asymmetrica’ in the text. GenBank numbers are listed in Supplementary Data Table S3.
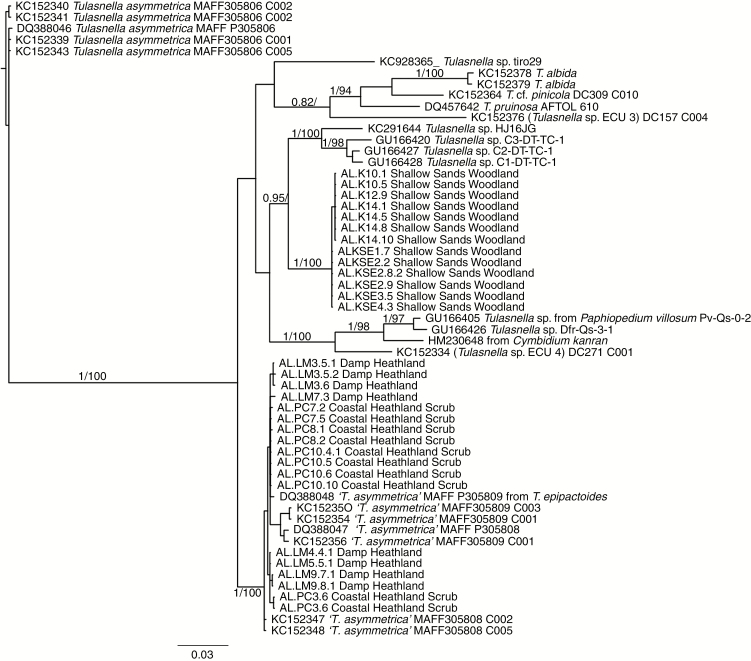
Fig. 4.
Concatenated phylogenetic tree for the ITS, nLSU1, nLSU2 and mtLSU region of Tulasnella isolates from the Shallow Sands Woodland, Damp Sands Heathland and Coastal Heathland sites. The sequences clustered into two well-supported (bootstrap = 100 %) OTUs (‘Tulasnella asymmetrica’ and OTU1), based on a 5 % sequence divergence cut-off (Linde et al., 2014, 2017). Warcup’s (1981) isolates of T. asymmetrica were separated into T. asymmetrica and ‘T. asymmetrica’ based on phylogenetic divergence (Cruz et al., 2014). We have kept this separation and refer to ‘T. asymmetrica’ as such in the text. Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.7sb22cb
Symbiotic germination: general methods
Seed collected from 7–10 hand-pollinated T. epipactoides individuals from each of the above three sites was combined and thoroughly mixed, air-dried and stored with dried silica crystals at 4 °C for up to 6 months for use in symbiotic seed germination tests. Seeds were surface-sterilized with 0.5 % NaOCl (10 % Domestos®) for 3 min, drained by vacuum onto a 3 μm pore filter, rinsed with sterile water and plated onto sterile filter paper on plates of oatmeal agar (OMA) (Clements and Ellyard, 1979) with 2 g L–1 sucrose and 0.1 g L–1 yeast extract in a laminar flow cabinet. A 1 cm OMA agar block colonized with an isolated fungus was placed on two ends of the filter paper. Plates were sealed with Parafilm® and incubated in the dark.
Symbiotic germination temperature trial
Sequencing results confirmed only two fungal OTUs, with only one OTU isolated at any given site. To test if temperature affected germination in OTU1 and ‘T. asymmetrica’, 27 fungal isolates (nine isolates from the SSW pertaining to OTU1, and 18 isolates pertaining to ‘T. asymmetrica’ which included nine isolates from each of the DH and CHS sites) were tested for symbiotic germination under three temperature cycles: 12 °C for 16 h–16 °C for 8 h, 16 °C for 16 h–24 °C for 8 h, or 27 °C constant. There were three replicated germination plates per fungal isolate per temperature treatment [thus a minimum of 27 replicates were tested of OTU1 (SSW) and 54 replicates for ‘T. asymmetrica’ (DH and CHS)]. Germination was scored as the presence or absence on the production of leaf primordia (Stage 5 of Warcup, 1981).
Symbiotic germination effect of fungal OTU on growth of adult plants
To test the effect of site of isolation of fungi and the effect of fungal OTU on growth of adult plants, germinated seedlings from the above trial were flasked using the methods of Reiter et al. (2016), and grown for a further 8 weeks before de-flasking. We de-flasked 35 T. epipactoides germinated with OTU1 from the SSW site and 70 T. epipactoides germinated with ‘T. asymmetrica’, 35 from each of the DH and CHS sites. Each seedling was de-flasked into Bio Gro Terrestrial Orchid Conservation Mix in 12 inch diameter plastic pots (one plant per pot) and watered in the glasshouse as required. We measured the diameter of the tuber of adult plants (12 months after deflasking) at the widest point as our growth measurement (plants lose there leaves annually so leaf measurements are not ideal). Thus symbiotically germinated T. epipactoides with fungi from the SSW as well as fungi from DH and CHS, were compared with a one-way analysis of variance (ANOVA) using the Analysis ToolPak, Microsoft Excel 2013. Potted plants were randomized in their positioning on benches throughout the glasshouse amongst the below co-planting trial and thus were also able to be used as controls in the co-planting experiment.
Co-planting with associated flora
We conducted a controlled glasshouse experiment with ten species of plant that were selected on the basis of co-occurring with T. epipactoides. The plants consisted of three species that naturally occurred at the SSW site, six species that naturally occurred at the DH and CHS sites, and one species, Rytidosperma caespitosum, that occurred at all sites. The number of co-occurring plants available to use of each species varied between 12 and 64 (Table 3). Seedlings from the above T. epipactoides germination trial germinated with either OTU1 (SSW) or ‘T. asymmetrica’ (DH and CHS) were used for the co-planting experiments. Thus each pot had one orchid seedling, grown with one fungal species, and one co-occurring plant species (Table 3). Plants naturally occurring at the SSW site were co-planted with T. epipactoides grown with OTU1, plants naturally occurring at the DH and CHS were co-planted with T. epipactoides grown with ‘T. asymmetrica’, and R. caespitosum which occurs across all three sites received in separate pots T. epipactoides grown with either OTU1 or ‘T. asymmetrica’. This experiment was conducted at the same time as the effect of fungal OTU experiment, allowing the potted plants grown with either OTU1 or ‘T. asymmetrica’ alone (without co-plants) to be used as controls.
Table 3.
Species co-planted with Thelymitra epipactoides
| Species co-planted with Thelymitra epipactoides | Shallow Sands Woodland | Damp Heathland | Coastal Heathland Scrub | OTU used | Replicates per OTU |
|---|---|---|---|---|---|
| Acacia euthycarpa (J.M.Black) J.M.Black | * | 1 | 12 | ||
| Allocasuarina verticillata (Lam.) L.A.S.Johnson | * | 1 | 26 | ||
| Rytidosperma caespitosuma | * | * | * | 1, ‘T. asymmetrica’ | 28, 64 |
| Callitris gracilis R.T.Baker | * | 1 | 12 | ||
| Kennedia prostrata R.Br. | * | ‘T. asymmetrica’ | 32 | ||
| Leptospermum continentale Joy Thomps. | * | * | ‘T. asymmetrica’ | 54 | |
| Lomandra longifolia Labill. | * | ‘T. asymmetrica’ | 32 | ||
| Poa poiformis (Labill.) Druce | * | ‘T. asymmetrica’ | 30 | ||
| Pultenaea canaliculata F.Muell. | * | ‘T. asymmetrica’ | 29 | ||
| Thomasia petalocalyx F.Muell. | * | ‘T. asymmetrica’ | 21 |
*Site of mycorrhizal isolates co-planted, OTU used and replicates per OTU. Includes nine isolates from the SSW relating to OTU1, and 18 isolates from the DH and CHS relating to ‘T. asymmetrica’.
aRytidosperma caespitosum (Gaudich.) Connor & Edgar was the only species to occur commonly across all three sites and was therefore the only species to be tested with both OTU1 and ‘T. asymmetrica’.
Potting was in winter while the orchid was actively growing. Seedlings were potted into Bio Gro Terrestrial Orchid Conservation Mix in 12 inch diameter plastic pots. Pots were randomized across glasshouse benches and incubated in a temperature-controlled glasshouse with 12 °C night and 25 °C day temperatures. Plants received natural light and were watered as required for 12 months. After 12 months of co-planting, tuber width (at the largest dimensions) of each orchid was measured. Results were analysed using a one-way ANOVA; subsequently a two-tailed t-test assuming unequal variance was performed to compare each pair of means (e.g. the respective T. epipactoides OTU without co-planted species control group mean compared with that of T. epipactoides OTU with specific co-planted species group mean) using the Analysis ToolPak, Microsoft Excel 2013.
RESULTS
Is there habitat-driven variation in Tulasnella associated with T. epipactoides?
Thirty-one Tulasnella isolates were successfully sequenced across all four regions. For the nLSU1 region, all fungi had amplicons of about 650 bp. For the nLSU2 region, all fungi had amplicons of about 350 bp. The DH and CHS isolates had mitochondrial amplicons about 100 bp larger than those from SSW. This was due to an insert at the 3’ end that was removed for the alignment and so had no influence on the relationships shown. Isolate ALPC3.4 was sequenced but not included in the trees because it had a short sequence (non-overlapping ITS1 and 4).
The ITS sequences formed two well-supported (bootstrap = 100 %) OTUs, based on a 5 % sequence divergence cut-off (Linde et al., 2014, 2017) (Fig. 3; Supplementary Data Table S2). A similar topology and clustering resulted for nLSU and mtLSU regions (data not shown). Consequently, a phylogenetic tree representing a concatenated data set is presented (Fig. 4). In all trees, sequences representing isolates from DH and CHS clustered together (Figs 3 and 4). Warcup’s (1981) isolates of T. asymmetrica were separated into T. asymmetrica and ‘T. asymmetrica’ based on phylogenetic divergence (Cruz et al., 2014). We have kept this separation and refer to ‘T. asymmetrica’ as such throughout this manuscript. Furthermore, these sequences [including seven reference sequences from GenBank, suggested as ‘T. asymmetrica’ (Cruz et al., 2014)], showed <3 % sequence divergence and therefore were considered ‘T. asymmetrica’ (Fig. 3; Supplementary Data Table S2). Sequences of isolates from SSW clustered into a unique, well-supported OTU (OTU1), with no close match to any GenBank sequence. All isolates from T. epipactoides clearly separated into two clades representing two fungal OTUs separated according to habitat, with isolates from the SSW pertaining to OTU1 and those from the DH and CHS pertaining to ‘T. asymmetrica’.
Do OTUs germinate orchid seed under different temperatures?
Symbiotic germination was successful at one or more temperatures with the majority of the 27 isolates tested. The number of plates showing germination was significantly affected by temperature (ANOVA, F = 6.76, P = 0.001). Tulasnella isolated from orchids in SSW (OTU1) germinated seed at the lower tested temperature cycles (12–16 °C and 16–24 °C) (Table 4). ‘Tulasnella asymmetrica’ from DH and CHS germinated seed at all three temperature ranges (Table 4).
Table 4.
Germination as a percentage of plates that germinated T. epipactoides seed to stage 5 with 27 isolates (nine from OTU1 and 18 from ‘T. asymmetrica’) under three temperatures
| Vegetation community | OTU | No. of isolates | Germination % | ||
|---|---|---|---|---|---|
| Low (12–16 °C) | Medium (16–24 °C) | High (27 °C) | |||
| Shallow Sands Woodland | 1 | 9 | 66.6 % (n = 27 plates) | 22.2 % (n = 27 plates) | 0 % (n = 27 plates) |
| Damp Heathland and Coastal Heathland Scrub | ‘T. asymmetrica’ | 18 | 77.7 % (n = 54 plates) | 83.3 % (n = 54 plates) | 66.6 % (n = 54 plates) |
Symbiotic germination effect of fungal OTU on growth of adult plants
We found no effect of fungal OTU on growth of adult plants. There was no significant difference in tuber size between mature orchids when comparing sites of OTU isolation (SSW, DH and CHS) on growth (ANOVA, F = 3.08, P = 0.652). There was no significant difference in tuber size between mature orchids grown with isolates of OTU1 (SSW) or ‘T. asymmetrica’ (DH and CHS) (ANOVA, F = 0.428, P = 0.652) when planted without associated flora.
Do symbiotically grown T. epipactoides plants benefit from co-planting with associated flora?
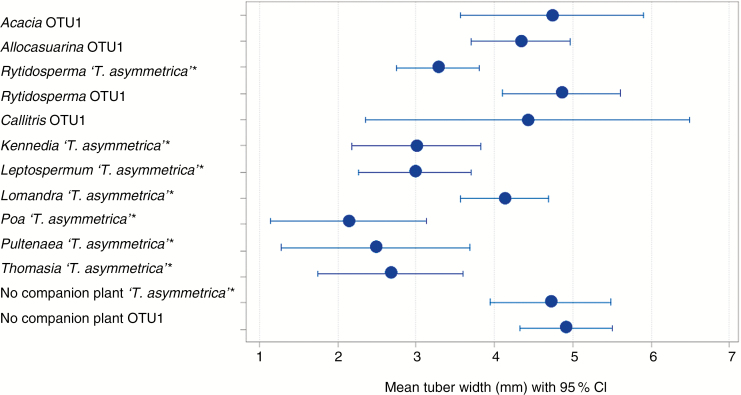
Tuber size of T. epipactoides co-planted with associated plant species was significantly different between treatments (ANOVA, F = 5.374, P < 0.001). Six associated species: Rytidosperma caespitosum, Kennedia prostrata, Pultenaea canaliculata, Poa poiformis, Leptospermum continentale and Thomasia petalocalyx co-planted with T. epipactoides grown with ‘T. asymmetrica’ isolates had a significant (P < 0.005) detrimental effect on the tuber size of T. epipactoides compared with those grown without companion plants (Supplementary Data Table S1). Co-planting with R. caespitosum resulted in significantly (P < 0.05) smaller tuber widths of T. epipactoides when orchid seed was grown with ‘T. asymmetrica’ compared with seed grown with OTU1 (Supplementary Data Table S1). Thelymitra epipactoides tuber size germinated with fungal OTU1 and subsequently grown by co-planting with associated plant species from the OTU1 site (SSW) (Fig. 5) did not differ significantly from control tubers grown without any co-plants.
Fig. 5.
Mean tuber width of seedlings of T. epipactoides grown with either fungus OTU1 or fungus ‘T. asymmetrica’, co-planted with co-occurring plant species after12 months in a pot trial against control without co-planted species. Lines represent the 95 % confidence interval calculated for each individual treatment. Significant differences (P ≤ 0.05) between control and co-occurring plant treatments are indicated by asterisks.
DISCUSSION
Highlights
This study provides clear evidence for two tulasnelloid OMFs associated with T. epipactoides. These two tulasnelloid fungi were not detected to co-occur at the same sites within pelotons isolated from T. epipactoides roots and were found in different habitats. Tulasnella OTU1 occurs in the drier SSW site while ‘T. asymmetrica’ occurs in the wetter DH and CHS sites. In addition, this is the first study to grow orchid seed to adult plants to test fitness differences with the use of different OMFs. However, the two OTUs tested did not result in differences in orchid tuber widths in adult plants. This study shows that the growth (measured by tuber width) of T. epipactoides germinated with OTU1 and ‘T. asymmetrica’, was significantly (P < 0.005) reduced when co-planted with five of ten tested co-occurring plant species compared with controls without co-occurring plant species. Furthermore, this study is the first to show that a terrestrial orchid has different optimal germination temperatures depending on the OMF species present.
Is there ecologically driven variation in Tulasnella associated with T. epipactoides?
Fungi isolated from T. epipactoides were tulasnelloid, as expected from a previous study on Thelymitra species in south-eastern Australia by Warcup (1981). Warcup (1981) isolated 79 Tulasnella strains from 15 of the then 35 Thelymitra species (Thelymitra was revised by Jeanes, 2002) showing that Thelymitra associated with seven species of Tulasnella, namely T. calospora, T. asymmetrica, T. cruciata, T. irregularis, T. violea, T. alantospora and an undescribed species.
The ITS region provides species delineation in Tulasnella that is equivalent to that from multiple nuclear and mitochondrial sequences (Cruz et al., 2011; Linde et al., 2014). The research presented here supports this conclusion, as sequences from the nLSU (28S) and mitochondrial ribosomes supported the same phylogenetic pattern as with the ITS region. All the Tulasnella fungi isolated in this study grouped into two distinct OTUs. One of the OTUs (OTU1) was only isolated from the SSW site, whereas isolates identified as ‘T. asymmetrica’ were obtained from the DH and CHS sites. Warcup’s (1981) isolates from Australian Thelymitra species named as T. asymmetrica, were separated into T. asymmetrica and ‘T. asymmetrica’ based on phylogenetic divergence (Cruz et al., 2014). We show that ‘T. asymmetrica’ and a new, but related lineage (OTU1), are associated with T. epipactoides. The percentage sequence divergence between ‘T. asymmetrica’ and OTU1 in the ITS region, exceeded a 5 % cut-off, as suggested from other Tulasnella studies (Linde et al., 2014, 2017). The maximum sequence divergence within each of OTU1 and ‘T. asymmetrica’ is <3 %, in line with the arbitrary figure of 97 % similarity for an OTU (McCormick et al., 2006; Nilsson et al., 2008; Peay et al., 2008) used by others (Roche et al., 2010; Jacquemyn et al., 2011; Kartzinel et al., 2013; Pandey et al., 2013).
The two Tulasnella symbionts found in this study did not co-occur at any of the three sites in the root samples, with OTU1 only detected in the SSW site (dry – forest), while ‘T. asymmetrica’ was found at sites DH and CHS (wetter – heathland). There are three possible explanations for the apparent habitat-driven switch of Tulasnella symbionts. The first is that a reduced suite of tulasnelloid fungi is present in each type of habitat; all may have been present initially but some may have declined due to intolerance of drier conditions or a decline in, or competition for, available resources for saprophytic growth (Brundrett et al., 2003; Mehra et al., 2017). This would suggest that fungal ecological specificity (Perkins and McGee, 1995) is the over-riding determinant of which fungi the orchid encounters and then orchid phylogeny determines whether or not a symbiosis is possible, rather than absolute specificity (Curtis, 1939). The second is that the entire suite of tulasnelloid fungi is present, but the orchid preferentially forms mycorrhizal symbioses with only one of the fungi depending on the environment, as shown in Goodyera pubescens (McCormick et al., 2006), which switch fungal associations in drought years. In other studies, Cypripedium acaule had a broad association between soil chemistry, elevation and root-associated fungi (Bunch et al., 2013) and Bipinnula species, which have soil nutrients modulating mycorrhizal associations (Mujica et al., 2016). A third possibility is that the orchid forms mycorrhizal associations predominantly with different fungi as it matures (McCormick et al., 2004; Otero et al., 2005; Bidartondo and Read, 2008). This is deemed unlikely as all orchids sampled were mature and seeds germinated equally with the fungi from mature plants from each site. All tested fungi were also able to maintain the orchids in pot trials to 18 months maturity. In other studies, with Tulasnella, Phillips et al. (2011) found the same fungi in mature orchids, seedlings and protocorms, whereas McCormick et al. (2004) found fungal diversity to be greater in adult plants than in protocorms.
Thelymitra epipactoides at the SSW site are exposed to lower rainfall, higher temperatures (Table 1; Fig. 1) and fewer nutrients (Grundy et al., 2015) than at the two mesic sites. Thelymitra epipactoides has a predominantly coastal distribution, and either expansion inland from the more mesic sites or remaining in these relict sites from a wetter time period may have been possible only because of the ability to form mycorrhiza with fungi that could also survive there, as discussed for tropical epiphytes (Martos et al., 2012). As conditions become less mesic (Hughes, 2003), it is likely that the range of fungi available to the orchid changes due to ‘environmental filtering’ (Kraft et al., 2015). Drought is suggested to have induced a switch in Tulasnella symbionts associated with Goodyera pubescens (McCormick et al., 2006). Kartzinel et al. (2013) found that a decrease in seasonal precipitation correlated with a decrease in diversity of the Tullasnellaceae in a tropical epiphytic orchid. Similarly, Phillips et al. (2016) found that OTUs of Sebacina associated with Caladenia stem-collars varied across soil types and rainfall in South West Australia Bio regions, with the Northern Wheatbelt only showing one Sebacina OTU and the Swan Coastal Plain (wetter than the Northern Wheatbelt) a larger diversity with six OTUs present (although sampling across each region was limited). Additionally, decreases in soil P and N decreased OTU richness from Bipinnula species in Chile (Mujica et al., 2016). The above findings suggest that some fungal OTUs are adapted to different environmental conditions. It is also possible that the mycorrhizal fungi are adapted to different habitats (Oja et al., 2015; Jacquemyn et al., 2016; Ruibal et al., 2017), though habitat and environmental conditions are difficult to tease apart. In the SSW habitat, which has more stressful environmental conditions (for fungi), it is likely that these limitations on available compatible fungi resulted in the orchids forming mycorrhiza with an available fungus different from that at the more mesic near-coastal sites.
The presence of two OTUs of Tulasnella forming effective mycorrhizal associations in climatically different sites of T. epipactoides is not surprising given the formerly widespread distribution of this endangered species. This is contrary to the hypothesis that threatened orchids have greater mycorrhizal specificity than common orchids and become rare because their compatible fungi are restricted geographically (Swarts et al., 2010). It also contrasts with high fungal specificity in all the Drakaea (Tulasnella secunda) (Phillips et al., 2011), Caleana (T. secunda) (Linde et al., 2014) and Chiloglottis (Tulasnella prima and T. sphagneti) species, which are mostly common (Linde et al., 2017). Furthermore, the rare Diuris fragrantissima was shown to associate with a range of closely related Tulasnella OTUs (Smith et al., 2010), and several OTUs have been found in some rare (Kartzinel et al., 2013; Pandey et al., 2013) and locally endemic orchids (Suárez et al., 2016). Therefore, we suggest that mycorrhizal specificity in T. epipactoides does not cause orchid rarity. The germination of Tipularia discolor only in the presence of wood colonized by specific fungi at a limited range of decomposition stages (McCormick et al., 2004) suggests that substrate and nutrition are important for determining the distribution of orchid mycorrhizae. It is thus feasible that fungal ecology (temperature, habitat, nutrient or pH requirements) may drive the differences in mycorrhizal fungi isolated from the three study sites of T. epipactoides.
Do Tulasnella OTUs have different temperature requirements for orchid germination?
The isolates from the hotter, drier SSW site (OTU1) supported germination only at cooler temperatures (12–16 °C and 16–24 °C) and not at all under the higher temperature conditions (27 °C). In contrast, ‘T. asymmetrica’, which was only present in the more mesic sites, supported germination at all tested temperatures (Table 4). These temperature ranges coincide best with temperatures when the majority of the precipitation is expected at each site. The SSW site is characterized by extreme temperatures regularly well in excess of 40 °C over the summer months, and is dry on average with only 400 mm of rainfall annually (Australian Government Bureau of Meteorology, 2017), with the greatest rainfall occurring in the cool wet winter. Thus, it is beneficial for orchid seed to germinate at winter temperatures, with seed germinated in summer unlikely to survive during the hot, dry summer conditions. At the mesic sites (DH and CHS), the cooler temperatures and greater coastal rainfall throughout the year may be more conducive to survival after germination, throughout the year.
Temperature can affect plant fungal symbiosis (Staddon et al., 2002; Kivlin et al., 2013) and has been shown to affect arbuscular mycorrhizal performance (Gavito et al., 2003). Seed from higher elevation locations often germinate at higher temperatures (Cavieres and Arroyo, 2000), though this is not consistent among species. Optimal germination temperatures may vary not only with altitude but also with other factors including light, seed batch and water (Giménez-Benavides et al., 2005; Bauk et al., 2016). While there are no orchid mycorrhizal relationships currently reported that are temperature dependent, this study suggests that high temperatures affect the ability of OTU1 to germinate T. epipactoides, with no germination seen at 27 °C.
The high (60–80 %) percentages of T. epipactoides plates that germinated with the optimal combination of fungi and temperature emphasizes that both OTUs were highly effective in germinating seed. Other germination trials with closely related tulasnelloid fungi isolated from Drakaea, Chiloglottis and Diuris species (Roche et al., 2010; Smith et al., 2010; Phillips et al., 2011) had relatively low germination success (42, 23 and 30 %, respectively). Those germination trials were only conducted at one constant temperature. It is possible that varying conditions might have increased germination, as suggested elsewhere (Phillips et al., 2011).
Do symbiotically grown T. epipactoides plants benefit from co-planting with associated flora?
There was no benefit from co-planting with any of the species that were tested, suggesting the absence of any benefit by providing a suitable microsite for the Tulasnella. The detrimental results due to co-planting with five of the ten co-planted species may be from restrictions in light and competition by the roots of the other species (Wilson, 1988; Calder et al., 1989), though competition between their associated micro-organisms cannot be excluded. Interestingly there was a significant (P < 0.005) difference in tuber width of T. epipactoides when grown with Rytidosperma caespitosum, with those germinated with OTU1 significantly (P < 0.05) larger than those germinated with ‘T. asymmetrica’, suggesting that ‘T. asymmetrica’ negatively affects T. epipactoides growth under those conditions. When grown without associated companion plants, orchids with different fungal isolates as inocula had the same tuber width. This suggests that fungal isolates belonging to the two OTUs do not differ in terms of the benefit they provide to the plant, except when grown with other plants. There are no other studies to our knowledge on long-term trials on the effect of fungal isolates on adult orchid plants. Such differences in growth caused by co-occurring plants may be examples of subtle interactions that affect long-term success in orchid conservation in the wild (Smith et al., 2010). Further investigation of the effects of different OTUs may support these observations.
Conservation implications
‘Tulasnella asymmetrica’ is globally distributed (Cruz et al., 2016), and many orchid mycorrhizal fungi are more widely distributed than the orchids with which they form symbiotic associations (Waud et al., 2017). Our results support previous research suggesting that mycorrhizal specificity is not the driving force for rarity in some orchids (Phillips et al., 2011; Kartzinel et al., 2013; Pandey et al., 2013).
There are three important conservation translocation considerations arising from this research: (1) with more than one species of mycorrhizal fungi able to germinate seed, it may be beneficial to introduce multiple fungal species in a conservation translocation; (2) alternatively, with differences in orchid germination temperatures between OMFs, the OMF has to be site matched to avoid recruitment losses; and (3) associated plants in a site may be detrimental to the growth of an orchid and need to be taken into consideration when selecting sites for conservation translocation at the microhabitat level.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: summary of significance values from two-sample t-tests. Table S2: pairwise minimum and maximum sequence divergences (percentage) between Tulasnella clades.
ACKNOWLEDGEMENTS
We thank the following people for assistance: Wendy Bedggood, Mary Argall, David Pitts, Ash Burns, Richard Thomson, the Australasian Native Orchid Society (Victorian Branch), the Horsham Secondary College students, and Ryan Phillips for providing feedback on a draft manuscript. This work was supported by the Australian Orchid Foundation (grant no. 273) and by National Landcare Programme funding through the Wimmera Catchment Management Authority. N.R. designed the research, collected and isolated the fungal isolates, germinated the seeds, grew the orchids in the pot trial and analysed the data; A.L. extracted and sequenced DNA; C.L. analysed sequencing data. All authors interpreted the data and wrote the paper. There was no involvement of the funding providers in designing the study; collecting, analysing or interpreting the data; or deciding to submit the paper for publication. The authors have no conflict of interest affecting this paper.
LITERATURE CITED
- Anderson SH, Kelly D, Ladley JJ, Molloy S, Terry J. 2011. Cascading effects of bird functional extinction reduce pollination and plant density. Science 331: 1068–1071. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bachman S, Barker A, et al. 2016. https://stateoftheworldsplants.com/2017/ State of the world’s plants – 2016.
- Australian Government Bureau of Meteorology 2017. Climate data online http://www.bom.gov.au/climate/data (accessed 26 November 2017).
- Backhouse G. 2007. Are our orchids safe down under? A national assessment of threatened orchids in Australia. Lankesteriana 7: 28–43. [Google Scholar]
- Batty A, Dixon K, Brundrett M, Sivasithamparam K. 2001. Constraints to symbiotic germination of terrestrial orchid seed in a mediterranean bushland. New Phytologist 152: 511–520. [DOI] [PubMed] [Google Scholar]
- Bauk K, Flores J, Ferrero C, Pérez-Sánchez R, Las Peñas ML, Gurvich DE. 2016. Germination characteristics of Gymnocalycium monvillei (Cactaceae) along its entire altitudinal range. Botany 95: 419–428. [Google Scholar]
- Bidartondo MI, Read DJ. 2008. Fungal specificity bottlenecks during orchid germination and development. Molecular Ecology 17: 3707–3716. [DOI] [PubMed] [Google Scholar]
- Brook BW, Sodhi NS, Bradshaw CJ. 2008. Synergies among extinction drivers under global change. Trends in Ecology and Evolution 23: 453–460. [DOI] [PubMed] [Google Scholar]
- Brundrett MC, Scade A, Batty AL, Dixon KW, Sivasithamparam K. 2003. Development of in situ and ex situ seed baiting techniques to detect mycorrhizal fungi from terrestrial orchid habitats. Mycological Research 107: 1210–1220. [DOI] [PubMed] [Google Scholar]
- Bunch WD, Cowden CC, Wurzburger N, Shefferson RP. 2013. Geography and soil chemistry drive the distribution of fungal associations in lady’s slipper orchid, Cypripedium acaule. Botany 91: 850–856. [Google Scholar]
- Calder D, Cropper S, Tonkinson D. 1989. The ecology of Thelymitra epipactoides F Muell (Orchidaceae) in Victoria, Australia, and the implications for management of the species. Australian Journal of Botany 37: 19–32. [Google Scholar]
- Cameron DD, Leake JR, Read DJ. 2006. Mutualistic mycorrhiza in orchids: evidence from plant–fungus carbon and nitrogen transfers in the green-leaved terrestrial orchid Goodyera repens. New Phytologist 171: 405–416. [DOI] [PubMed] [Google Scholar]
- Cavieres LA, Arroyo MT. 2000. Seed germination response to cold stratification period and thermal regime in Phacelia secunda (Hydrophyllaceae) – altitudinal variation in the Mediterranean Andes of central Chile. Plant Ecology 149: 1–8. [Google Scholar]
- Clements MA, Ellyard RK, 1979. The symbiotic germination of Australian terrestrial orchids. American Orchid Society Bulletin 48: 810–816. [Google Scholar]
- Clements MA, Muir H, Cribb PJ. 1986. A preliminary report on the symbiotic germination of European terrestrial orchids. Kew Bulletin 41: 437–445. [Google Scholar]
- Coates F, Jeanes J, Pritchard A. 2002. National Recovery Plan for twenty-five threatened orchid taxa of Victoria, South Australia and New South Wales 2003–2007. Department of Sustainability and Environment, Victoria. [Google Scholar]
- Cropper S, Calder D. 1990. The floral biology of Thelymitra epipactoides (Orchidaceae), and the implications of pollination by deceit on the survival of this rare orchid. Plant Systematics and Evolution 170: 11–27. [Google Scholar]
- Cruz D, Suárez JP, Kottke I, Piepenbring M, Oberwinkler F. 2011. Defining species in Tulasnella by correlating morphology and nrDNA ITS-5.8 S sequence data of basidiomata from a tropical Andean forest. Mycological Progress 10: 229–238. [Google Scholar]
- Cruz D, Suárez JP, Kottke I, Piepenbring M. 2014. Cryptic species revealed by molecular phylogenetic analysis of sequences obtained from basidiomata of Tulasnella. Mycologia 106: 708–722. [DOI] [PubMed] [Google Scholar]
- Cruz D, Suárez JP, Piepenbring M. 2016. Morphological revision of Tulasnellaceae, with two new species of Tulasnella and new records of Tulasnella spp. for Ecuador. Nova Hedwigia 102: 279–338. [Google Scholar]
- Curtis JT. 1939. The relation of specificity of orchid mycorrhizal fungi to the problem of symbiosis. American Journal of Botany 26: 390–399. [Google Scholar]
- Davis BJ, Phillips RD, Wright M, Linde CC, Dixon KW. 2015. Continent-wide distribution in mycorrhizal fungi: implications for the biogeography of specialized orchids. Annals of Botany 116: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearnaley JDW, Martos F, Selosse MA. 2012. Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects. In: Hock B, ed. Fungal associations. Berlin Heidelberg: Springer, 207–230. [Google Scholar]
- Fochi V, Chitarra W, Kohler A, et al. . 2017. Fungal and plant gene expression in the Tulasnella calospora–Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytologist 213: 365–379. [DOI] [PubMed] [Google Scholar]
- Gavito ME, Schweiger P, Jakobsen I, 2003. P uptake by arbuscular mycorrhizal hyphae: effect of soil temperature and atmospheric CO2 enrichment. Global Change Biology 9: 106–116. [Google Scholar]
- Giménez-Benavides L, Escudero A, Pérez-García F. 2005. Seed germination of high mountain Mediterranean species: altitudinal, interpopulation and interannual variability. Ecological Research 20: 433–444. [Google Scholar]
- Grundy M, Rossel RV, Searle R, Wilson P, Chen C, Gregory L. 2015. Soil and landscape grid of Australia. Soil Research 53: 835–844. [Google Scholar]
- Harcourt AH, Coppeto S, Parks S. 2002. Rarity, specialization and extinction in primates. Journal of Biogeography 29: 445–456. [Google Scholar]
- Hughes L. 2003. Climate change and Australia: trends, projections and impacts. Austral Ecology 28: 423–443. [Google Scholar]
- Huynh TT, Thomson R, Mclean CB, and Lawrie AC. 2009. Functional and genetic diversity of mycorrhizal fungi from single plants of Caladenia formosa (Orchidaceae). Annals of Botany 104: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCNS 2013. Guidelines for reintroductions and other conservation translocations. Gland, Switzerland: IUCN Species Survival Commission. [Google Scholar]
- Jacquemyn H, Merckx V, Brys R, Tyteca D, Cammue B, Honnay O, Lievens B. 2011. Analysis of network architecture reveals phylogenetic constraints on mycorrhizal specificity in the genus Orchis (Orchidaceae). New Phytologist 192: 518–528. [DOI] [PubMed] [Google Scholar]
- Jacquemyn H, Deja A, Bailarote BC, Lievens B. 2012. Variation in mycorrhizal associations with tulasnelloid fungi among populations of five Dactylorhiza species. PLoS One 7: e42212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemyn H, Waud M, Lievens B, and Brys R. 2016. Differences in mycorrhizal communities between Epipactis palustris, E. helleborine and its presumed sister species E. neerlandica. Annals of Botany 118: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanes JA. 2002. A taxonomic revision of the genus Thelymitra J. and G. Forst. (Orchidaceae) in Australia. Journal of the Native Orchid Society of South Australia 26: 52. [Google Scholar]
- Jeanes JA. 2011. Resolution of the Thelymitra aristata (Orchidaceae) complex of south-eastern Australia. Muelleria 29: 110–129. [Google Scholar]
- Jeanes JA. 2013. An overview of the Thelymitra nuda (Orchidaceae) complex in Australia including the description of six new species. Muelleria 31: 3–30. [Google Scholar]
- Jeanes JA, Backhouse G. 2006. Wild orchids of Victoria, Australia. Australia: Aquatic Photographics. [Google Scholar]
- Johnson S, Steiner K. 1997. Long-tongued fly pollination and evolution of floral spur length in the Disa draconis complex (Orchidaceae). Evolution 51: 45–53. [DOI] [PubMed] [Google Scholar]
- Kartzinel TR, Trapnell DW, Shefferson RP. 2013. Highly diverse and spatially heterogeneous mycorrhizal symbiosis in a rare epiphyte is unrelated to broad biogeographic or environmental features. Molecular Ecology 22: 5949–5961. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivlin SN, Emery SM, Rudgers JA. 2013. Fungal symbionts alter plant responses to global change. American Journal of Botany 100: 1445–1457. [DOI] [PubMed] [Google Scholar]
- Kraft NJ, Adler PB, Godoy O, James EC, Fuller S, Levine JM. 2015. Community assembly, coexistence and the environmental filtering metaphor. Functional Ecology 29: 592–599. [Google Scholar]
- Linde CC, Phillips RD, Crisp MD, Peakall R. 2014. Congruent species delineation of Tulasnella using multiple loci and methods. New Phytologist 201: 6–12. [DOI] [PubMed] [Google Scholar]
- Linde CC, May TW, Phillips RD, Ruibal M, Smith LM, Peakall R. 2017. New species of Tulasnella associated with terrestrial orchids in Australia. IMA FUNGUS 8: 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA. 2000. Biotic invasions: causes, epidemiology, global consequences, and control. Ecological Applications 10: 689–710. [Google Scholar]
- Martos F, Munoz F, Pailler T, Kottke I, Gonneau C, Selosse MA. 2012. The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Molecular Ecology 21: 5098–5109. [DOI] [PubMed] [Google Scholar]
- McCormick MK, Jacquemyn H. 2014. What constrains the distribution of orchid populations?New Phytologist 202: 392–400. [Google Scholar]
- McCormick M, Whigham D, O’Neill J. 2004. Mycorrhizal diversity in photosynthetic terrestrial orchids. New Phytologist 163: 425–438. [DOI] [PubMed] [Google Scholar]
- McCormick MK, Whigham DF, Sloan D, O’Malley K, Hodkinson B. 2006. Orchid–fungus fidelity: a marriage meant to last?Ecology 87: 903–911. [DOI] [PubMed] [Google Scholar]
- Mehra S, Morrison PD, Coates F, Lawrie AC. 2017. Differences in carbon source utilisation by orchid mycorrhizal fungi from common and endangered species of Caladenia (Orchidaceae). Mycorrhiza 27: 95–108. [DOI] [PubMed] [Google Scholar]
- Mujica MI, Saez N, Cisternas M, Manzano M, Armesto JJ, Pérez F. 2016. Relationship between soil nutrients and mycorrhizal associations of two Bipinnula species (Orchidaceae) from central Chile. Annals of Botany 118: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. 2008. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolutionary Bioinformatics 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oja J, Kohout P, Tedersoo L, Kull T, Kõljalg U. 2015. Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytologist 205: 1608–1618. [DOI] [PubMed] [Google Scholar]
- Orejuela-Gartner JE. 2012. Orchids of the cloud forests of southwestern Colombia and opportunities for their conservation. European Journal of Environmental Sciences 2: 19–32. [Google Scholar]
- Otero JT, Bayman P, Ackerman JD. 2005. Variation in mycorrhizal performance in the epiphytic orchid Tolumnia variegata in vitro: the potential for natural selection. Evolutionary Ecology 19: 29–43. [Google Scholar]
- Pandey M, Sharma J, Taylor D, Yadon VL. 2013. A narrowly endemic photosynthetic orchid is non-specific in its mycorrhizal associations. Molecular Ecology 22: 2341–2354. [DOI] [PubMed] [Google Scholar]
- Pauw A, Hawkins JA. 2011. Reconstruction of historical pollination rates reveals linked declines of pollinators and plants. Oikos 120: 344–349. [Google Scholar]
- Peay KG, Kennedy PG, Bruns TD. 2008. Fungal community ecology: a hybrid beast with a molecular master. Bioscience 58: 799–810. [Google Scholar]
- Perkins A, McGee P. 1995. Distribution of the orchid mycorrhizal fungus, Rhizoctonia solani, in relation to its host, Pterostylis acuminata, in the field. Australian Journal of Botany 43: 565–575. [Google Scholar]
- Phillips RD, Barrett MD, Dixon KW, Hopper SD. 2011. Do mycorrhizal symbioses cause rarity in orchids?Journal of Ecology 99: 858–869. [Google Scholar]
- Phillips RD, Peakall R, Hutchinson MF, et al. . 2014. Specialized ecological interactions and plant species rarity: the role of pollinators and mycorrhizal fungi across multiple spatial scales. Biological Conservation 169: 285–295. [Google Scholar]
- Phillips RD, Barrett MD, Dalziell EL, Dixon KW, Swarts ND. 2016. Geographical range and host breadth of Sebacina orchid mycorrhizal fungi associating with Caladenia in south-western Australia. Botanical Journal of the Linnean Society 182: 140–51. [Google Scholar]
- Pimm SL, Jenkins CN, Abell R, et al. . 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science 344: 1246752. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. 2012. FigTree. Version 1.4. 0 Available at http://tree.bio.ed.ac.uk/software/figtree/
- Rasmussen HN. 1990. Cell differentiation and mycorrhizal infection in Dactylorhiza majalis (Rchb. f.) Hunt & Summerh. (Orchidaceae) during germination in vitro. New Phytologist 116: 137–147. [Google Scholar]
- Rasmussen HN. 1995. Terrestrial orchids: from seed to mycotrophic plant.Cambridge: Cambridge University Press. [Google Scholar]
- Rasmussen HN. 2002. Recent developments in the study of orchid mycorrhiza. Plant and Soil 244: 149–163. [Google Scholar]
- Reiter N, Whitfield J, Pollard G, et al. . 2016. Orchid re-introductions: an evaluation of success and ecological considerations using key comparative studies from Australia. Plant Ecology 217: 81–95. [Google Scholar]
- Reiter N, Vlcek K, O’Brien N, et al. . 2017. Pollinator rarity limits reintroduction sites in an endangered sexually deceptive orchid (Caladenia hastata): implications for plants with specialized pollination systems. Botanical Journal of the Linnean Society 184: 122–136. [Google Scholar]
- Robertson AW, Kelly D, Ladley JJ, Sparrow AD. 1999. Effects of pollinator loss on endemic New Zealand mistletoes (Loranthaceae). Conservation Biology 13: 499–508. [Google Scholar]
- Roche SA, Carter RJ, Peakall R, Smith LM, Whitehead MR, Linde CC. 2010. A narrow group of monophyletic Tulasnella (Tulasnellaceae) symbiont lineages are associated with multiple species of Chiloglottis (Orchidaceae): implications for orchid diversity. American Journal of Botany 97: 1313–1327. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, et al. . 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruibal MP, Triponez Y, Smith LM, Peakall R, Linde CC. 2017. Population structure of an orchid mycorrhizal fungus with genus-wide specificity. Scientific Reports 7: 5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse MA, Martos F. 2014. Do chlorophyllous orchids heterotrophically use mycorrhizal fungal carbon?Trends in Plan Science 19: 683–685. [DOI] [PubMed] [Google Scholar]
- Selosse MA, Roy M. 2009. Green plants that feed on fungi: facts and questions about mixotrophy. Trends in Plant Science 14: 64–70. [DOI] [PubMed] [Google Scholar]
- Shefferson RP, Weiß M, Kull T, Taylor D. 2005. High specificity generally characterizes mycorrhizal association in rare lady’s slipper orchids, genus Cypripedium. Molecular Ecology 14: 613–626. [DOI] [PubMed] [Google Scholar]
- Smith ZF, James EA, McLean CB. 2010. Mycorrhizal specificity of Diuris fragrantissima (Orchidaceae) and persistence in a reintroduced population. Australian Journal of Botany 58: 97–106. [Google Scholar]
- Staddon P, Heinemeyer A, Fitter A. 2002. Mycorrhizas and global environmental change: research at different scales. Plant and Soil 244: 253–261. [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57: 758–771. [DOI] [PubMed] [Google Scholar]
- Stöckel M, Těšitelová T, Jersáková J, Bidartondo MI, Gebauer G. 2014. Carbon and nitrogen gain during the growth of orchid seedlings in nature. New Phytologist 202: 606–615. [DOI] [PubMed] [Google Scholar]
- Suárez JP, Eguiguren JS, Herrera P, Jost L. 2016. Do mycorrhizal fungi drive speciation in Teagueia (Orchidaceae) in the upper Pastaza watershed of Ecuador?Symbiosis 69: 161–168. [Google Scholar]
- Swarts ND, Dixon KW. 2009. Terrestrial orchid conservation in the age of extinction. Annals of Botany 104: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts ND, Sinclair EA, Francis A, Dixon KW. 2010. Ecological specialization in mycorrhizal symbiosis leads to rarity in an endangered orchid. Molecular Ecology 19: 3226–3242. [DOI] [PubMed] [Google Scholar]
- Taylor D, Bruns T, Leake J, Read D. 2002. Mycorrhizal specificity and function in myco-heterotrophic plants. Mycorrhizal Ecology 157: 375–413. [Google Scholar]
- Thomas CD, Cameron A, Green RE, et al. . 2004. Extinction risk from climate change. Nature 427: 145–148. [DOI] [PubMed] [Google Scholar]
- Tilman D, May RM, Lehman CL, Nowak MA. 1994. Habitat destruction and the extinction debt. Nature 371: 65–66. [Google Scholar]
- Vallee L. 2004. Guidelines for the translocation of threatened plants in Australia. Canberra, Australia: Australian Network for Plant Conservation. [Google Scholar]
- Warcup J. 1971. Specificity of mycorrhizal association in some Australian terrestrial orchids. New Phytologist 70: 41–46. [Google Scholar]
- Warcup JH. 1981. The mycorrhizal relationships of Australian orchids. New Phytologist 87: 371–381. [Google Scholar]
- Waterman RJ, Bidartondo MI. 2008. Deception above, deception below: linking pollination and mycorrhizal biology of orchids. Journal of Experimental Botany 59: 1085–1096. [DOI] [PubMed] [Google Scholar]
- Waud M, Brys R, Van Landuyt W, Lievens B, Jacquemyn H. 2017. Mycorrhizal specificity does not limit the distribution of an endangered orchid species. Molecular Ecology 26: 1687–1701. [DOI] [PubMed] [Google Scholar]
- Weiss M, Selosse MA, Rexer KH, Urban A, Oberwinkler F. 2004. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycological Research 108: 1003–1010. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor JL. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications. New York: Academic Press, 315–322. [Google Scholar]
- Wilson JB. 1988. Shoot competition and root competition. Journal of Applied Ecology 25: 279–296. [Google Scholar]
- World Checklist of Selected Plant Families 2017. Facilitated by the Royal Botanic Gardens, Kew Available at: http://wcsp.science.kew.org (accessed 24 November 2017)
- Zubek S, Majewska ML, Błaszkowski J, Stefanowicz AM, Nobis M, Kapusta P. 2016. Invasive plants affect arbuscular mycorrhizal fungi abundance and species richness as well as the performance of native plants grown in invaded soils. Biology and Fertility of Soils 52: 879–893. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.