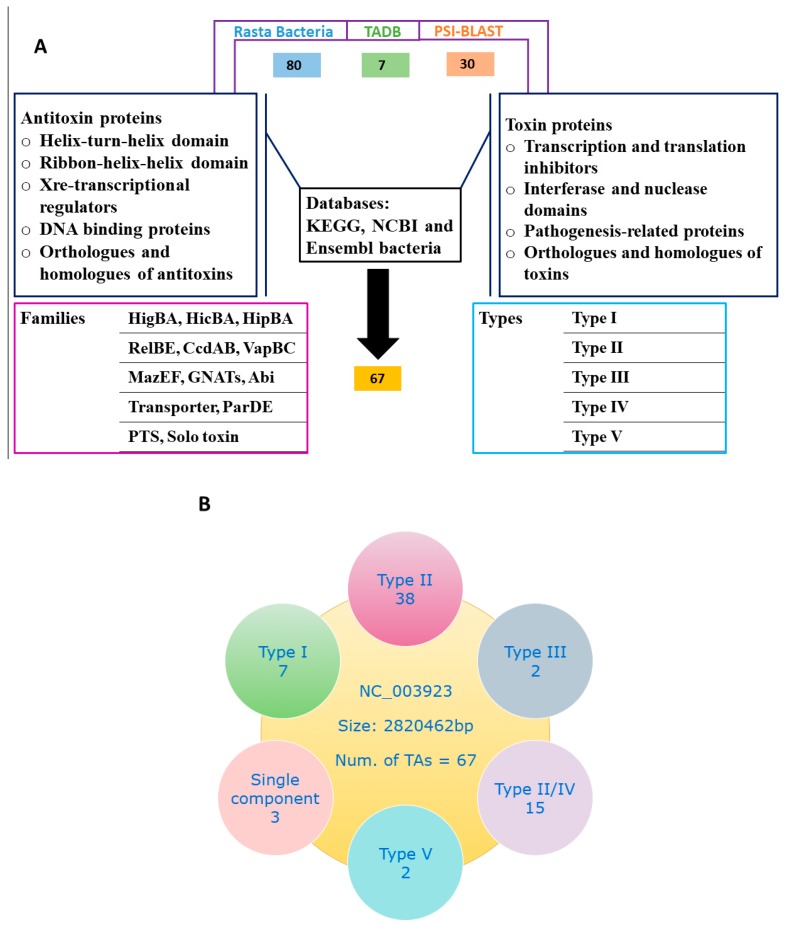
Figure 1.
Screening of TA systems. (A) Preliminary screening of TA loci in S. aureus genome was performed with Rasta bacteria and toxin-antitoxin database (TADB) and followed by position-specific iterative basic local alignment search tool (PSI-BLAST) searches. In total, 80 putative TA systems are predicted by Rasta bacteria, seven by TADB, and 30 by PSI-BLAST. TA systems were manually curated by conserved domains such as helix-turn-helix (HTH), ribbon-helix-helix (RHH) and transcriptional regulators for antitoxin, and conserved domains such as interferases, nucleases, transcription and translation inhibitors and pathogenesis-related proteins for toxins. All selected TA system orthologues and homologs were analyzed, and finally, 67 putative TA systems were selected. (B) The selected 67 TA systems were classified into different families and types. Type II TA systems were abundantly detected followed by putative type II/IV TA systems.