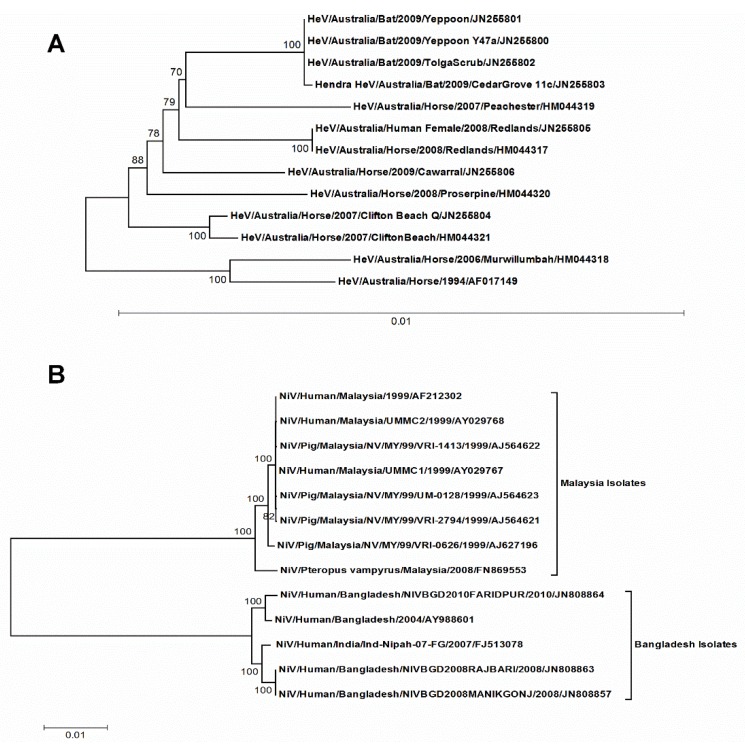
Figure 1.
The maximum likelihood trees of HeV (A) and NiV (B) complete coding genomic sequences. The trees were constructed by TN93 + G substitution model implemented in MEGA software version 7. The reliability of the trees was assessed by bootstrap with 1000 replications. The bootstrap values greater than 70 are shown. Scale bars represent substitutions per site. For each strain, the following data are furnished: Virus/Species affected/Country/strain name/year of isolation/GenBank accession nos. HeV, Hendra virus; NiV, Nipah virus.