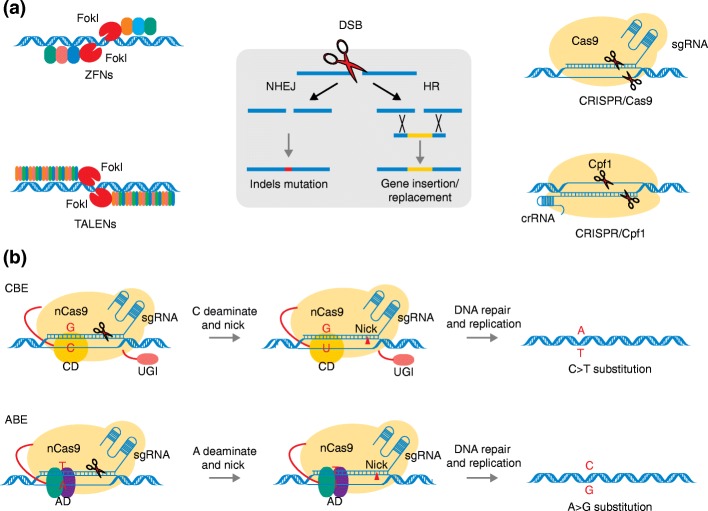
Fig. 1.
a Genome editing tools and DNA repair mechanisms. ZFNs and TALENs on the left panel use FokI endonuclease to cut DNA double strands. Since FokI functions as a dimer, when two ZFNs or TALENs bind their targets and bring the FokI monomers into close proximity, cleavage occurs. CRISPR/Cas9 system on the right panel employs sgRNA for DNA binding and Cas9 protein for DNA cleavage. While CRISPR/Cpf1 system uses crRNA for DNA binding and Cpf1 protein for DNA cleavage. On the middle panel, when DSB was produced by genome editing techniques, the plant’s endogenous repair systems fix the DSB by NHEJ or HR. NHEJ introduces small indels (red line) into the DSB and results in frame-shift mutations or premature stop codons. HR can cause gene replacements and insertions (yellow line) in the presence of a homologous donor DNA spanning the DSB. b Illustration of CRISPR/Cas9-mediated base editing. In the CBE system, nCas9 was fused to CD and UGI, and this complex could convert cytosine (C) in the targeting region to uracil (U), then U is changed to thymine (T) in DNA repair or replication processes, creating a C•G to T•A substitution. In the ABE system, nCas9 was fused to AD, and this system converts adenine (A) in the targeting region to inosine (I), which is treated as guanine (G) by polymerases, creating A•T to G•C substitutions. ABE adenine deaminases-mediated base editing, AD adenine deaminases, CBE cytidine deaminase-mediated base editing, CD cytidine deaminases, CRISPR clustered regularly interspaced short palindromic repeats, crRNA CRISPR RNA, DSB double-strand break, HR homologous recombination, nCas9 Cas9 nickase, NHEJ non-homologous end joining, sgRNA single-guide RNA, TALEN transcription activator-like effector nuclease, UGI uracil glycosylase inhibitor, ZFN zinc-finger nuclease