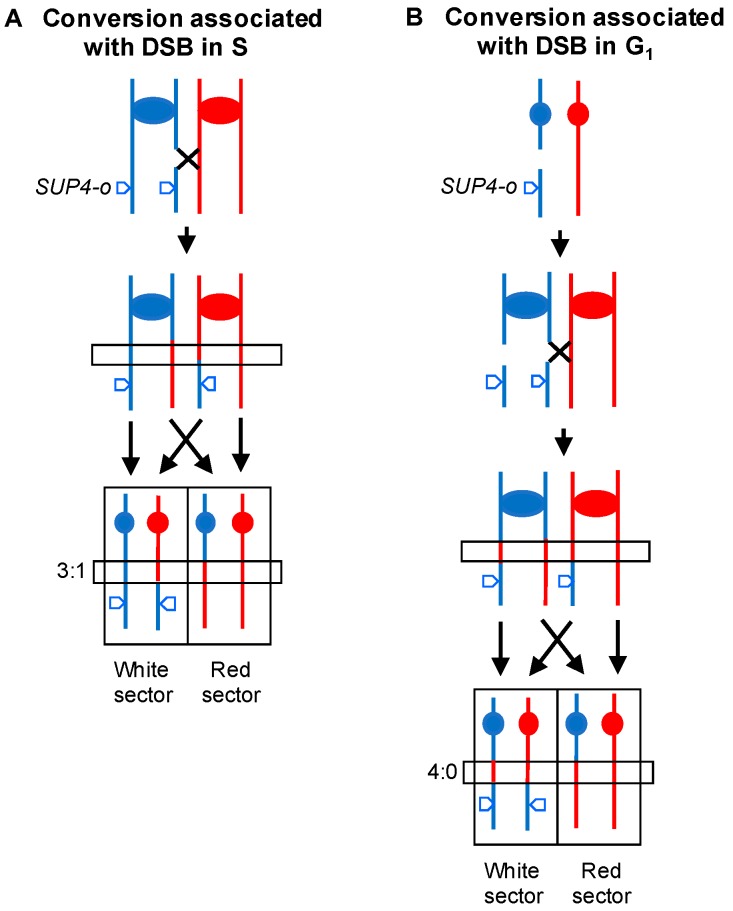
Figure 3.
Detecting gene conversion events associated with crossovers initiated in G2 or G1. A diploid is constructed that is heterozygous for an insertion of the SUP4-o ochre suppressing tRNA gene, and homozygous for the ade2-1 ochre-suppressible allele. The diploid forms pink colonies, but derivatives that have zero or two copies of SUP4-o result in red or white colonies, respectively. Thus, crossovers can produce a red/white sectored colony. These sectors are analyzed by SNP arrays to determine the location of the recombination breakpoints. The region of conversion (boxed in the figure) is detected as a difference in the location of the breakpoints in the two sectors. (A) Crossover as a consequence of a DSB on one chromatid. As a consequence of the conversion associated with repair of a single broken chromatid, a 3:1 conversion (a region in which 3 of the chromatids contain sequences from the red homolog and one contains sequences from the blue homolog) is observed. (B) Crossover reflecting the repair of two DSBs. We show a broken chromosome that is replicated to yield two chromatids broken at the same position. The conversion tracts associated with these repair events result in a 4:0 region. To explain the pattern of LOH, we suggest that one break is repaired to generate a crossover, and the second is repaired to produce a conversion event unassociated with a crossover.