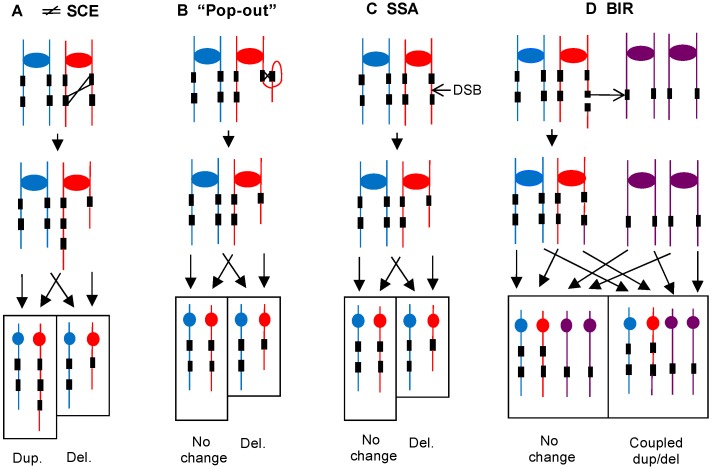
Figure 6.
Homologous recombination between non-allelic repeats as a mechanism for generating large deletions and duplications. Blue and red lines show the two homologs of the diploid, and the purple lines indicate a different homolog. Black rectangles depict directly repeated genes. Arrows near the bottom of each panel show patterns of segregation with rectangles outlining the two daughter cells. Duplicated and unduplicated centromeres are shown as ovals and circles, respectively. (A) Unequal sister-chromatid exchange. Unequal crossing over between non-allelic repeats will produce one chromosome with a deletion and one with a duplication. Such unequal crossovers could also occur between homologs. (B) “Pop-out” event. An intrachromatid crossover between direct repeats will produce a chromosome with a deletion and a circular DNA molecule. The circle would likely be lost because it lacks a centromere. (C) Single-strand annealing (SSA). In this mechanism, a DSB occurs between two directly oriented repeats. Following processing of the broken ends, the repeats can anneal, resulting in loss of one repeat and the intervening DNA sequences [22]. (D) BIR. A DSB occurs in a repeat in one of the red chromatids with loss of the terminal fragment. The centromere-containing broken end invades a repeat on a non-homolog (purple chromatids) and copies the sequences from the point of invasion to the chromosome terminus. The resulting event generates a translocation. The daughter cell with the translocation will contain a large terminal deletion of “red” sequences and a large terminal duplication of “purple” sequences.