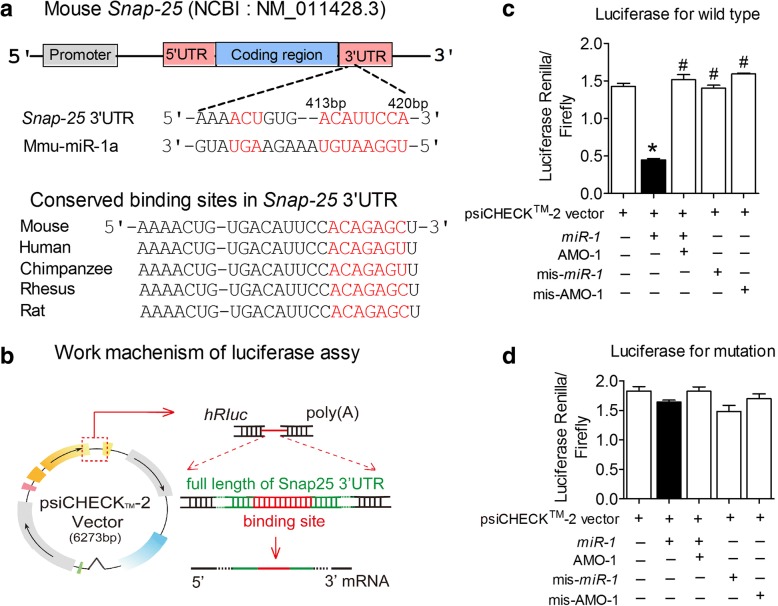
Fig. 3.
Snap25 is a potential target of miR-1. a Complementarity between the miR-1 seed sequence (5’end 2 ~ 8 nucleotides) and the 3’UTR of mouse’s Snap25 gene as predicted by a computational and bioinformatics-based approach using the Targetscan 5.1 algorithm. Watson-Crick complementarity is marked by a red colour. b The mechanism of the luciferase assay. The full-length 3’UTR of Snap25 was amplified by PCR and cloned into the psiCHECK™-2-control vector. c-d Luciferase reporter gene assay for interactions between miR-1 and its binding site (c) and the mutated binding site (d) in the 3’UTR of the Snap25 gene in HEK293T cells. HEK293T cells were transfected with psiCHECK™-2 vector, miR-1 mimics, AMO-1, or negative control siRNAs (mis-miR-1 and mis-AMO-1) using Lipofectamine 2000 (Wild type: PLevene = 0.086, one-way ANOVA: F = 122.906, P < 0.0001; 3’UTR mut: PLevene = 0.658, One-way ANOVA: F = 3.382, P = 0.054). *P < 0.05 versus psiCHECK™-2-control vector; #P < 0.05, versus miR-1, n = 3 batches of cells for each group