Abstract
Background
Biting midges of the genus Culicoides (Diptera: Ceratopogonidae) are small hematophagous insects responsible for the transmission of bluetongue virus, Schmallenberg virus and African horse sickness virus to wild and domestic ruminants and equids. Outbreaks of these viruses have caused economic damage within the European Union. The spatio-temporal distribution of biting midges is a key factor in identifying areas with the potential for disease spread. The aim of this study was to identify and map areas of neglectable adult activity for each month in an average year. Average monthly risk maps can be used as a tool when allocating resources for surveillance and control programs within Europe.
Methods
We modelled the occurrence of C. imicola and the Obsoletus and Pulicaris ensembles using existing entomological surveillance data from Spain, France, Germany, Switzerland, Austria, Denmark, Sweden, Norway and Poland. The monthly probability of each vector species and ensembles being present in Europe based on climatic and environmental input variables was estimated with the machine learning technique Random Forest. Subsequently, the monthly probability was classified into three classes: Absence, Presence and Uncertain status. These three classes are useful for mapping areas of no risk, areas of high-risk targeted for animal movement restrictions, and areas with an uncertain status that need active entomological surveillance to determine whether or not vectors are present.
Results
The distribution of Culicoides species ensembles were in agreement with their previously reported distribution in Europe. The Random Forest models were very accurate in predicting the probability of presence for C. imicola (mean AUC = 0.95), less accurate for the Obsoletus ensemble (mean AUC = 0.84), while the lowest accuracy was found for the Pulicaris ensemble (mean AUC = 0.71). The most important environmental variables in the models were related to temperature and precipitation for all three groups.
Conclusions
The duration periods with low or null adult activity can be derived from the associated monthly distribution maps, and it was also possible to identify and map areas with uncertain predictions. In the absence of ongoing vector surveillance, these maps can be used by veterinary authorities to classify areas as likely vector-free or as likely risk areas from southern Spain to northern Sweden with acceptable precision. The maps can also focus costly entomological surveillance to seasons and areas where the predictions and vector-free status remain uncertain.
Electronic supplementary material
The online version of this article (10.1186/s13071-018-3182-0) contains supplementary material, which is available to authorized users.
Keywords: Culicoides, Random Forest, Machine Learning, Europe, Monthly distribution, Spatial distribution, Presence-absence data, Targeted surveillance
Background
Culicoides (Diptera: Ceratopogonidae) biting midges are small blood-sucking insects responsible for the transmission of viruses causing the European outbreaks of bluetongue (BT) and Schmallenberg diseases in wild and domestic ruminant livestock [1, 2], and for African horse sickness in equids [1, 3]. BTV historically made sporadic incursions into some countries of the Mediterranean Basin (Portugal, Spain, the Greek islands close to Turkey and Cyprus) but from 1998 onwards the situation worsened when five other serotypes spread within France (Corsica), Italy, Greece and countries in the Balkans region [4]. BT was never reported in northern Europe until August 2006, when an unprecedented bluetongue virus (BTV) serotype 8 outbreak started in the border region of Germany, Belgium and the Netherlands and, over the next two years, it spread further over central and northern Europe [5–8]. This epidemic had a significant economic impact within the European Union, as a consequence of the restriction of animal movements and the large amount of financial resources invested in vaccination campaigns and vector surveillance programs [9–11]. In northern Europe, the Afro-Asian vector Culicoides imicola Kieffer is absent and therefore, the vector species incriminated in the transmission of BTV were the Palaearctic species belonging to the Obsoletus ensemble Culicoides obsoletus (Meigen)/Culicoides scoticus Downes & Kettle [12, 13], Culicoides chiopterus (Meigen) [14, 15] and Culicoides dewulfi Goetghebuer [16].
Many factors contribute to the transmission of vector-borne diseases, including the presence of infected hosts, competent vectors and suitable environmental temperatures for the pathogen to replicate inside the vector [17]. In the absence of ongoing entomological surveillance, a temporal map of the potential distribution of the vectors is key for health authorities to quickly delimitate possible areas and time periods of risk for disease transmission in the case of an outbreak of a known or emerging vector-borne disease [18–20]. The spatial distribution and phenology of vectors can be predicted from climate and environmental variables such as temperature, precipitation and land cover [18]. Temporal occurrence data (the presence or absence of a species at a specific time) in non-sampled areas or periods can be modelled using statistical techniques. This methodology is used to generate species distribution maps depicting the probability of the species being present at a given time [21], thus identifying areas with low or null adult activity and therefore, periods during which animal movements are safe.
Since the start of the BT outbreaks, European authorities have established a series of regulations for BT surveillance including vector monitoring to analyse the seasonal fluctuation of the vector populations and determine the seasonal vector-free periods (SVFP) for different regions [22, 23]. The EU defines SVFP by using a threshold on the abundance of female specimens, considering the parity stage of the Culicoides caught in the traps. This approach has been used to estimate the SVPF in Scotland for species of the Obsoletus group [24]. The authors estimated phenological events for each species such as the start and end of the SVFP. Brugger et al. [23] estimated vector-free periods in Austria using an approach based on the European Commission definition but without considering parity stage of female specimens. In the present study, we identified months where adult activity is null or very low, based on the monthly mean abundance for each farm, without considering the parity of the specimens collected as previously proposed by the EU legislation. Our definition of adult activity is different but comparable to the vector-free season defined by this legislation and, therefore, we keep the term “vector-free” season or period to refer to a period of the year with neglectable adult activity.
The SVFP during the winter was not ubiquitous across all European countries. Austria [23], Switzerland [25] and Sweden [26] reported the existence of a SVFP, while other countries such as Germany, France, Belgium and the Netherlands reported that a SVFP might not exist in these countries [16, 27–29]. Imposing restrictions of animal movement in areas where the vector is not present has a negative economic impact as the restriction is unnecessary. On the other hand, allowing animal movement in areas where the vector is present poses a risk of spreading infections to new areas, if environmental conditions are suitable for the virus to develop inside the vector. Being able to define vector-free areas and periods is not only useful for BT management, but also for emerging Culicoides-borne diseases in the future. For instance, Schmallenberg virus appeared suddenly in 2011 in Germany, and spread throughout 29 European countries [30], causing economic losses for sheep and cattle farmers [31]. In addition, the spread of African horse sickness has previously been reported in horses in Spain in 1966 and Spain and Portugal from 1987 to 1990 [32]. Knowing the geographical distribution of vectors allows veterinary authorities to focus control measurements in those areas at a specific time of year.
In this study, we used entomological data of C. imicola, Obsoletus ensemble and Pulicaris ensemble collected from nine European countries over a seven-year period. This entomological dataset was used previously to analyse the temporal fluctuation at different latitude bands for Europe, to analyse the start of the season at the geographical NUTS level and to interpolate the observed Culicoides abundance spatially [32]. In this work, we use the machine learning algorithm “Random Forest” (RF) to model the average monthly presence/absence observed and predict the probability of presence of C. imicola, Obsoletus ensemble and Pulicaris ensemble in unsampled areas, using climatic and environmental variables as predictors. The aim of this work was to predict areas and months likely to be free of biting midges or likely to have vectors as well as areas of uncertain status that need to be targeted for entomological surveillance in case of an outbreak. The resulting maps represent the first spatial distribution model for a transect comprising nine European countries from southern Spain to northern Sweden. The maps are useful tools as inputs for decision making by veterinary authorities to detect areas with adult activity and use this information to focus financial resources for active entomological surveillance programs.
Methods
Culicoides data
We used entomological data collected in farms from Spain, France, Germany, Switzerland, Austria, Denmark, Sweden, Norway and Poland between 2007 and 2013 as part of national surveillance programs or research projects [33]. For each trap site, observations consisted of the number of C. imicola, Obsoletus ensemble [C. obsoletus, C. scoticus, Culicoides montanus Shakirzjanova, Culicoides chiopterus (Meigen) and C. dewulfi] and Pulicaris ensemble [Culicoides pulicaris (Linnaeus) and Culicoides punctatus (Meigen)]. Culicoides biting midges were sampled from a total of 904 livestock farms comprising 31,429 trap collections. Onderstepoort traps were used for sampling biting midges, except for Germany (Biogents Sentinel traps) and in Spain (mini CDC traps). For these two countries, we multiplied the number of Culicoides for each observation by a conversion factor to make the number of specimens comparable between the different trapping methods. Details of both the sample protocols and the conversion factors used have been published previously [33].
For C. imicola and each of the Culicoides ensembles, we split the observation data set into 12 subsets according to month of the year. For each 12 monthly dataset, we calculated the average abundance on each farm for each year sampled. This resulted in 12 datasets with farms containing one monthly average abundance per year sampled. Then, we classified each monthly average each year into Presence or Absence according to the average abundance of the vector. Based on the European Union regulation [22] for the definition of the SVFP, in which an abundance threshold of biting midges is proposed to define Presence or Absence, we considered each monthly average for each year as Presence when it was above or equal to an abundance threshold of five midges for the Obsoletus and Pulicaris ensembles, and one specimen for C. imicola. Even though the European Union definition of Presence is based on the catch of five parous specimens per observation, we here considered the number of midges without differentiating females into their gonotrophic stage because this information was missing for some of the countries. This will result in a more conservative definition of SVFP. Our approach also differed from the approach used by the EU commission as for each farm we only classified the monthly average each year into Presence or Absence, and not each of the individual observations (when there were several observations per month).
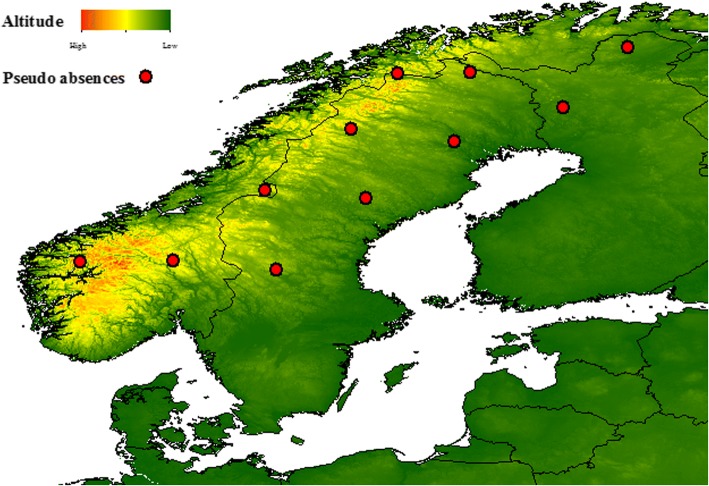
We constructed preliminary Random Forest (RF) models using occurrence data from January and February. The data collected in this period did not include any farms from northern Scandinavia. The resulting models predicted the occurrence of biting midges in January and February in this region (data not shown). However, earlier studies have reported an absence of biting midges in the Scandinavian peninsula during winter [26, 34]. Therefore, it was useful to provide pseudo-absence points to the models in order to increase their accuracy for predicting absences in the area. For January and February, we created 11 random pseudo-absence points above 60 degrees latitude in the highlands in Norway, central and northern Sweden and Finland and were added by hand using ArcMap 10.1 (ESRI, Redlands, CA, USA) (Fig. 1).
Fig. 1.
Eleven pseudo-absence points added to Norway, Sweden and Finland for January and February
Predictor variables
We used raster files (images) of 112 environmental and climatic variables, land cover and livestock density, each with a 1 km2 spatial resolution.
The environmental predictors included Mid-infrared (MIR), daytime Land Surface Temperature (dLST), nighttime Land Surface Temperature (nLST), Enhanced Vegetation Index (EVI) and Normalized Difference Vegetation Index (NDVI) as predictor variables. Each variable was derived from a MODIS temporal series from 2001 to 2012, and subjected to Temporal Fourier Analysis (FTA) [35]. For each environmental variable, the resulting products of FTA were the 14 images described in Table 1. This dataset was originally created by the TALA research group at the Department of Zoology at Oxford University, and was provided through the EDENext project [36].
Table 1.
Products of Temporal Fourier Analysis obtained from a single variable
| Fourier component | Description |
|---|---|
| A0 | Fourier mean for the entire time series |
| A1 | Amplitude of annual cycle |
| A2 | Amplitude of bi-annual cycle |
| A3 | Amplitude of tri-annual cycle |
| P1 | Phase of annual cycle |
| P2 | Phase of bi-annual cycle |
| P3 | Phase of tri-annual cycle |
| DA | Proportion of total variance due to all three cycles |
| D1 | Proportion of total variance due to annual cycle |
| D2 | Proportion of total variance due to bi-annual cycle |
| D3 | Proportion of total variance due to tri-annual cycle |
| MN | Minimum value |
| MX | Maximum value |
| VR | Total variance |
Each product corresponds to a raster image (1 km2 resolution) derived from a single environmental variable (for instance, NDVI)
We also included WORLDCLIM altitude data (digital elevation model) and bioclimatic variables as climatic predictors for Culicoides distribution. BIOCLIM images were obtained from the WORLDCLIM database [37, 38] (Table 2).
Table 2.
MODIS Fourier-transformed, BIOCLIM and Corine Land Cover predictors used to model the probability of Culicoides presence
| Source | Code | Description |
|---|---|---|
| MODIS (Fourier transformed) 2001–2012 | MIR | Mid-infrared |
| dLST | Daytime land surface temperature | |
| nLST | Nighttime land surface temperature | |
| NDVI | Normalized difference vegetation index | |
| EVI | Enhanced vegetation index | |
| BIOCLIM 1960–1990 | BIO 1 | Annual mean temperature |
| BIO 2 | Mean diurnal range: mean of monthly (max. temp - min. temp) | |
| BIO 3 | Isothermality (BIO2/BIO7) (×100) | |
| BIO 4 | Temperature seasonality (standard deviation × 100) | |
| BIO 5 | Maximum temperature of warmest month | |
| BIO 6 | Minimum temperature of coldest month | |
| BIO 7 | Temperature annual range (BIO5-BIO6) | |
| BIO 8 | Mean temperature of wettest quarter | |
| BIO 9 | Mean temperature of driest quarter | |
| BIO 10 | Mean temperature of warmest quarter | |
| BIO 11 | Mean temperature of coldest quarter | |
| BIO 12 | Annual precipitation | |
| BIO 13 | Precipitation of wettest month | |
| BIO 14 | Precipitation of driest month | |
| BIO 15 | Precipitation seasonality (coefficient of variation) | |
| BIO 16 | Precipitation of wettest quarter | |
| BIO 17 | Precipitation of driest quarter | |
| BIO 18 | Precipitation of warmest quarter | |
| BIO 19 | Precipitation of coldest quarter | |
| Altitude | Digital elevation model (DEM) | |
| Corine Land Covera | CLC 12 | Non-irrigated arable land |
| CLC 13 | Permanently irrigated land | |
| CLC 15–17 | Vineyards, fruit trees and berry plantations, olive groves | |
| CLC 18 | Pastures | |
| CLC 19 | Annual crops associated with permanent crops | |
| CLC 20 | Complex cultivation patterns | |
| CLC 21 | Land principally occupied by agriculture with significant areas of natural vegetation | |
| CLC 22 | Agro-forestry areas | |
| CLC 23 | Broad-leaved forest | |
| CLC 24 | Coniferous forest | |
| CLC 25 | Mixed forest | |
| CLC 26 | Natural grasslands | |
| CLC 29 | Transitional woodland-shrub | |
| CLC 35 | Inland marshes | |
| CLC 40 | Water courses | |
| CLC 41 | Water bodies |
aCLC plus the number refers to the CORINE land cover class used for modelling
We used a Corine Land Cover (CLC) map with 250 m pixel resolution to extract information on 16 relevant land cover classes (Table 2). For each class, we created a binary image with pixel values of 1 and 0 according the presence or absence of the class. Due to the higher spatial resolution of the CLC map compared to the other predictors, we resampled each of the binary class images to a resolution of 1 km2. This was done by overlaying a grid with cells of 1 km2 resolution. To each of these cells, we assigned the sum of all pixels with a value of 1 within them. Each 1 km2 cell of the grid was made up of 16 (4 × 4) pixels of the original CLC map. This resulted in new images for each land cover class with a pixel resolution of 1 km2, representing the frequency of each of the 16 different classes found in every 1 km2 area (pixel) on a scale of 0–16. CLC map was obtained from the European Environment Agency website [39].
We obtained livestock density data for cattle, goats, sheep, small ruminants and chickens from the Food and Agriculture Organization repository “GeoNetwork”. This dataset consisted of a series of raster files with information regarding livestock density at a global scale (“The gridded livestock of the world”) [40].
Modelling the probability of presence
Combining our Culicoides data with the predictors, we explored modelling approaches using VECMAP© software, v.2.0.16350.2473. For the final modelling of each month and each species, we used the Random Forest (RF) machine learning technique [41, 42] in R v.3.4.2 [43] (packages caret [44] and randomForest [45]) to model the probability of presence (PP) in the nine European countries using the Presence/Absence observations calculated at each farm. For each month we obtained a map showing the PP at the same resolution as the predictors (1 km2). The RF algorithm consists of an ensemble of decision trees used to predict the probability of class membership where the response variable is categorical (e.g. classification into presence and absence). An advantage of RF is the model’s capability of detecting nonlinear relationships between the response and the predictor variables [46] and that RF can handle a large number of predictor variables [46]. In addition, RF can produce a list of the most important predictors and scale them from 0–100 according to their importance as calculated by permuting each predictor and measuring the prediction error after the permutation [44].
The number of farms sampled varied from month to month. As expected, during summer more farms were sampled compared to winter, as in many countries of northern Europe entomological surveillance is not carried out during the cold winter months. For each monthly dataset, we used a stratified random split to divide the data into two subsets: one included 70% of the farms containing at least one year classified as presence together with the farms with only absence observations (training set). The second subset contained the remaining 30% of the farms as a test set to evaluate model performance [42, 47, 48]. We conducted a stratified random split based on farm ID in order to avoid having observations belonging to the same farm in both the training and the evaluation datasets (Table 3).
Table 3.
Total number of farms sampled each month and number of farms in the training and test sets
| Month | Total no. of sampled farms | Training set (70%) | Test set (30%) |
|---|---|---|---|
| January | 444 | 310 | 134 |
| February | 457 | 319 | 138 |
| March | 473 | 331 | 142 |
| April | 522 | 364 | 158 |
| May | 527 | 368 | 159 |
| June | 518 | 362 | 156 |
| July | 581 | 406 | 175 |
| August | 636 | 445 | 191 |
| September | 620 | 433 | 187 |
| October | 522 | 365 | 157 |
| November | 500 | 349 | 151 |
| December | 448 | 313 | 135 |
All observations belonging to a single farm were included in either the training or test set, but never in both
The number of Culicoides caught per farm highly varied between the different years. In this work, we considered each farm’s monthly classification into Presence or Absence for each year and included them in the training set as independent observations. Therefore, a farm might contain Presence and Absence observations from different years depending on the variation in mean monthly abundance between the different years.
The monthly Presence/Absence data were highly imbalanced, meaning that it contained a high proportion of one of the classes (Presence or Absence), i.e. the majority class. We investigated and compared five different balancing methods (no balancing, down-sampling, oversampling, ROSE [49], SMOTE [50], Tomek [50]) to cope with the imbalance and to improve model performance. We ran cross-validation (CV) for each balancing method 10 times with different random seeds and the best method was chosen according to highest AUC (data not shown). The balancing method chosen to balance the training set was oversampling, which entails duplicating the observations for the minority class in order to reach the same number of observations as the majority class [42]. We used the balanced training set of each month to train the RF model, and used the test sets to calculate the receiver operating characteristics (ROC) curve [42, 51, 52] and the area under this curve (AUC). We used the AUC as a measurement of model performance. AUC values close to 0.5 indicate that the model is not able to classify new samples better than random, values between 0.7 and 0.8 indicate acceptable model performance, values from 0.8 to 0.9 indicate excellent performance and values above 0.9 are considered outstanding [53]. For each month, we performed 5-fold CV to optimize the model parameter “mtry” (i.e. number of predictors used at each split). The “ntrees” parameter (number of trees of the forest) was set to 1000 trees in all cases.
For C. imicola, after the test set was created, we removed all the observations from farms not belonging to Spain or France, as the vector was not found in the seven remaining countries [33]. This reduced the large amount of Absence observations in the test set, which have an influence in the distribution of the classes.
Classification
Classification of predicted probabilities into Presence/Absence classes can be determined using a predetermined threshold (in ecology studies, normally the default is a PP of 0.5 [54]). Here, we were interested in defining a data-dependent threshold, as a predefined threshold of 0.5 might not be optimal [54]. The monthly PP maps obtained from our RF models were classified into three categories. We calculated a lower and upper threshold and all areas with a PP below the lower threshold were considered to be in the Absence class, while the areas with a PP above the upper threshold were classified as Presence areas. Regions with a PP between the two thresholds could not be classified as either Absence or Presence class, and were therefore classified as an Uncertain status category that may be targeted for active vector surveillance. The Absence and Presence classes refer here to the occurrence of adult activity and not to the ecological establishment of the vector, as in the classical species distribution modelling.
Lower and upper thresholds were calculated using the density function for the PP predicted by the model for each test set class (true presence/absence). To define the two thresholds for each month, we derived two gain functions Gpresence, Gabsence for 100 possible thresholds from 0 to 1, based on the area under the density function for Presence and Absence, respectively. We calculated Gpresence as the probability of a true presence and subtracted the probability of a misclassified presence multiplied by a parameter δ, which indicates the cost of a misclassified presence relative to a true presence. Similarly, we calculated Gabsence as the probability of a corrected classified absence (true absence) and subtracted the probability of misclassified absence multiplied by parameter γ, which indicates the cost of a misclassified absence relative to a true absence. Setting δ = 2, for example, means that the cost of a false positive classification is twice the gain of a true positive classification. The gain value can be considered in terms of timely initiation of countermeasures and a lower probability of an epidemic and trade restrictions, while the loss value would be the cost to the farm and society of incorrectly applied countermeasures. Similarly, for the interpretation of γ, the gain of a true negative classification and the loss from a false negative classification can be likened to being declared free from disease, with the cost to both farmer and society of a subsequent discovery of the disease. Similar considerations can be used to relate δ and γ to each other. If, for example, we set δ = ρ * γ in Eq. 1, the cost of misclassifying a presence is ρ times the cost of misclassifying an absence. We assign δ = 2 * γ in order to assign twice the importance to the Presence misclassifications compared to Absence misclassifications and we set γ = 2 to still give some importance to the Absences misclassifications.
The equations for Gpresence, Gabsence were:
| 1 |
| 2 |
where q represents the possible threshold value between 0 to 1, and where δ and γ are loss parameters.
To calculate the lower threshold, we used Eq. 1 to find the optimal upper threshold when assuming a loss parameter of δ = 4 by optimizing the gain Gpresence. Similarly, Eq. 2 was used to find the optimal lower threshold, assuming a loss parameter γ = 2. The upper and lower thresholds depend on the predictive power of the model, being more separated when the overlapping between classes is large. If the model performance is good, the overlapping between classes will be less and the two thresholds will be closer together.
In order to evaluate the sensitivity of the thresholds to the distribution of different test sets, we divided each monthly test set into ten equally sized folds (10 subsets) and calculated the density functions using nine out of the ten folds. This procedure was repeated for all the different folds (10 times), excluding a different fold each time, and plotted the new lower and upper thresholds together in the same graph. We applied this 10-fold cross-validation scheme to compare the threshold calculated with different subsets of the test set versus the thresholds calculated using all the observations of the test set.
We classified the monthly probability maps into the three classes: “Absence”, “Uncertain” and “Presence” using the thresholds calculated from all the observations of the test set.
Results
Obsoletus ensemble
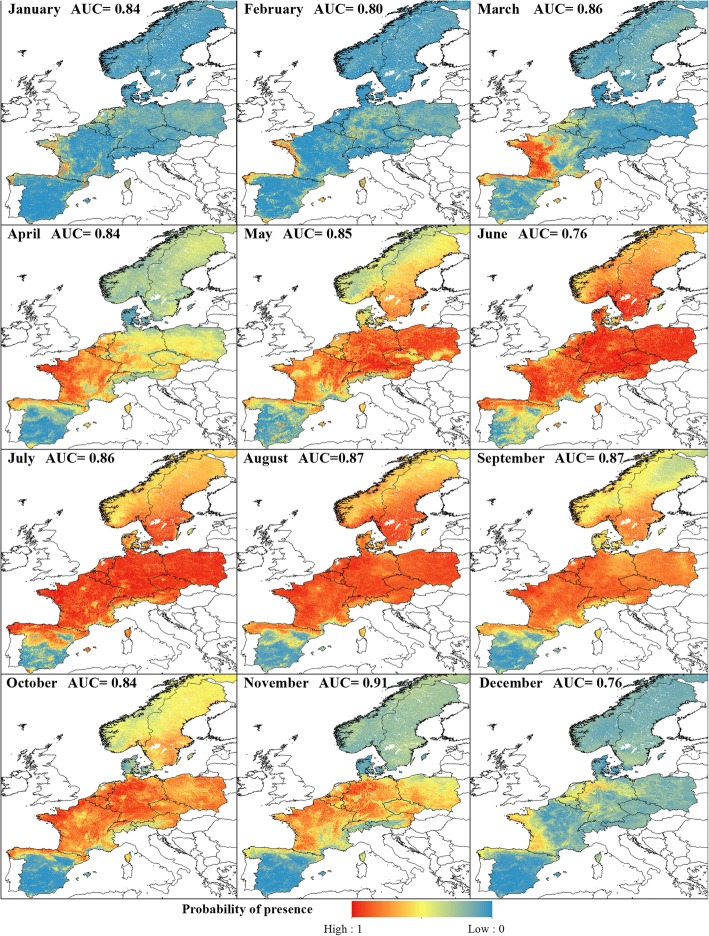
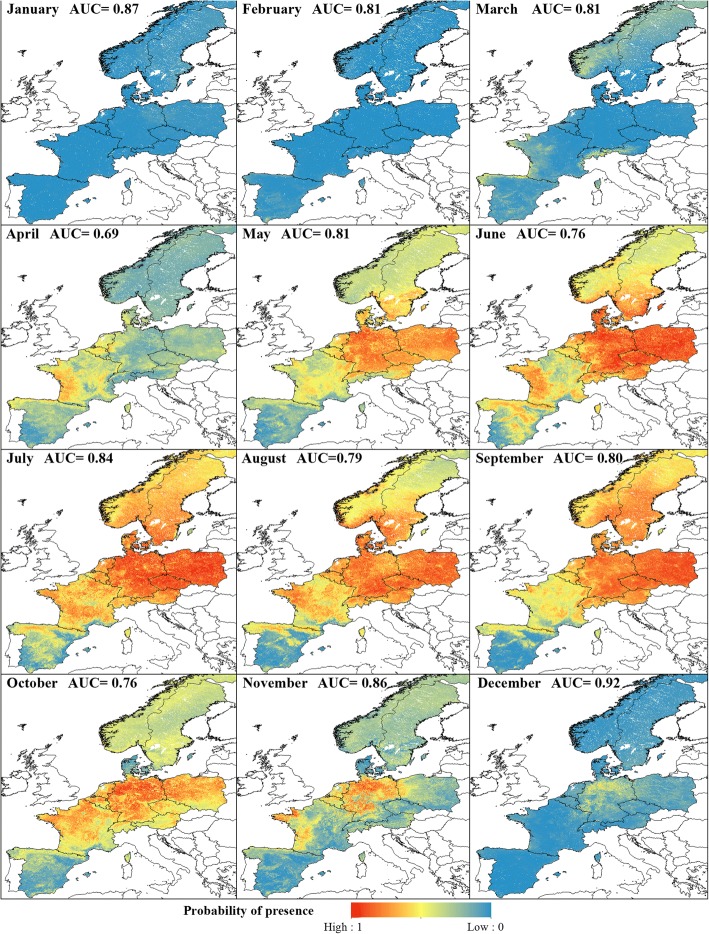
The 12 models were shown to perform well for the Obsoletus ensemble, with an AUC ranging from 0.76 in June and December to 0.91 in November (mean AUC = 0.84) (Fig. 2).
Fig. 2.
Predicted monthly probability of presence of Obsoletus ensemble. Monthly model performance is shown as the AUC value
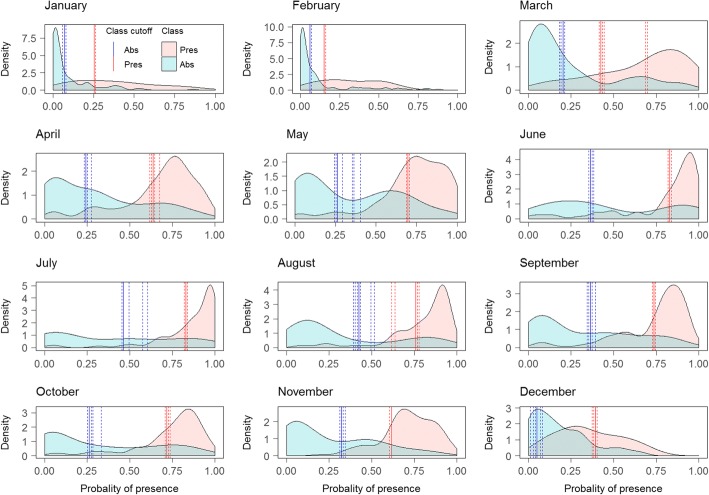
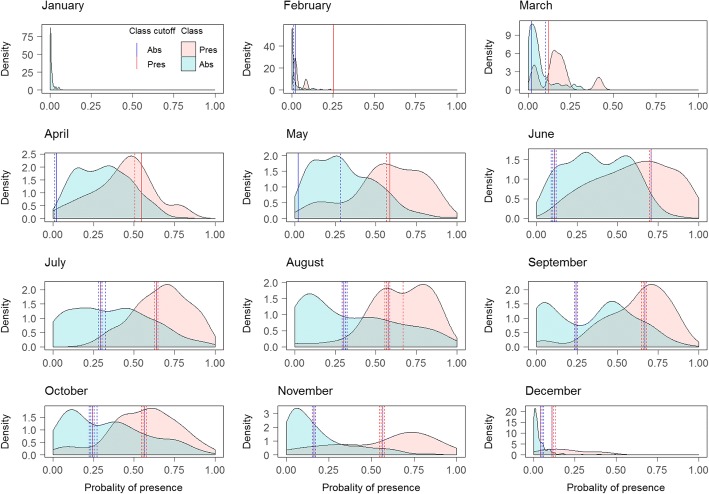
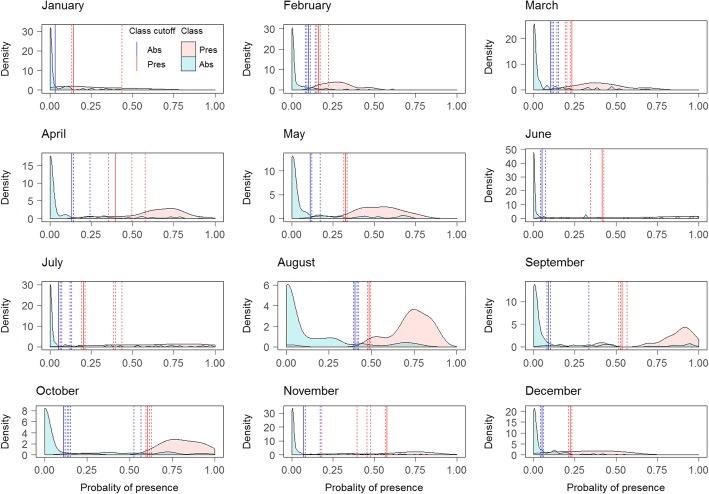
The majority class shifted from Absence in December-March, to Presence in April-November, and the models generally had good predictive power when predicting the majority class. However, the models performed less well when predicting the minority class. For January and February, the model predicted the Presence class relatively poorly, with a relatively flat density function (Fig. 3). The additional thresholds calculated using 10-fold CV were similar to the main threshold, indicating that the distribution of classes in the test set were robust when subtracting 10% of the data. The lower thresholds showed more variation compared to the variation of the upper thresholds (Fig. 3).
Fig. 3.
Obsoletus ensemble: monthly distribution of Presence and Absence classes of the test set samples as a function of their predicted probability of presence. Dashed lines show the additional thresholds calculated from 10-fold CV
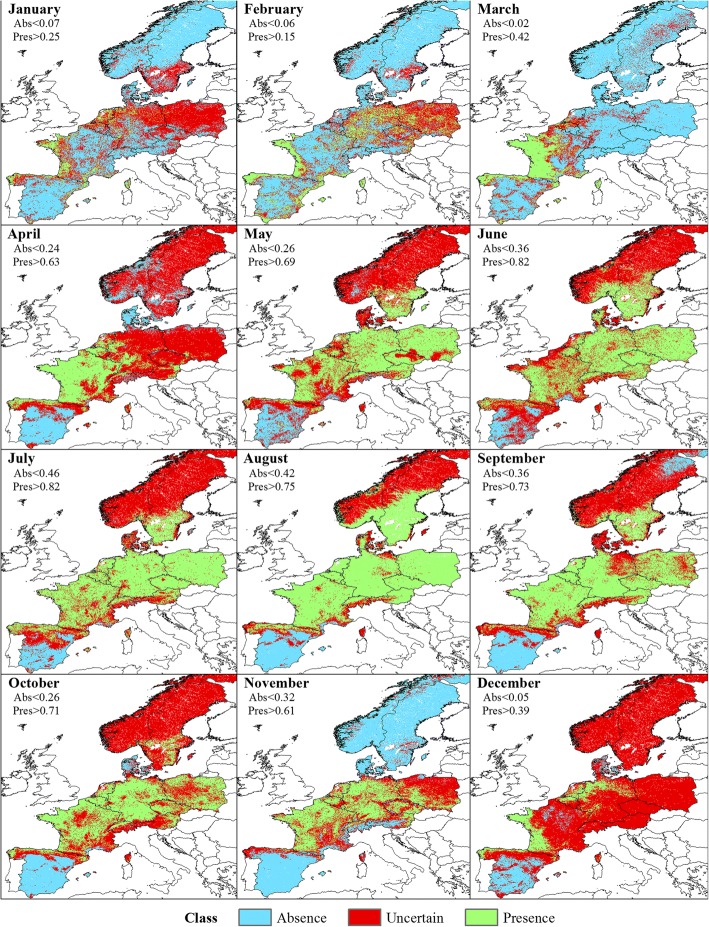
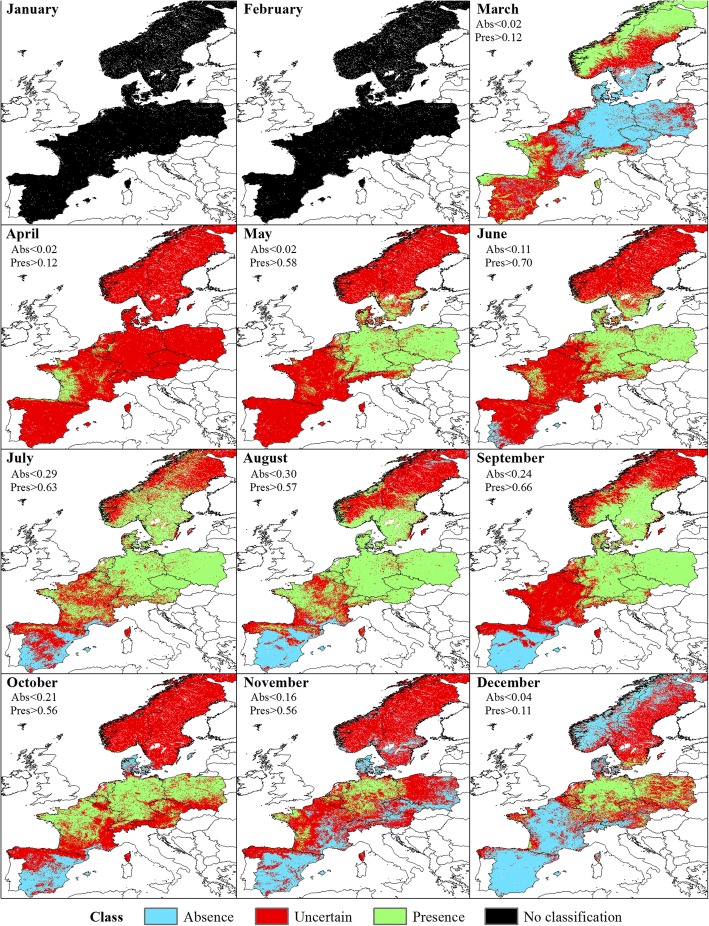
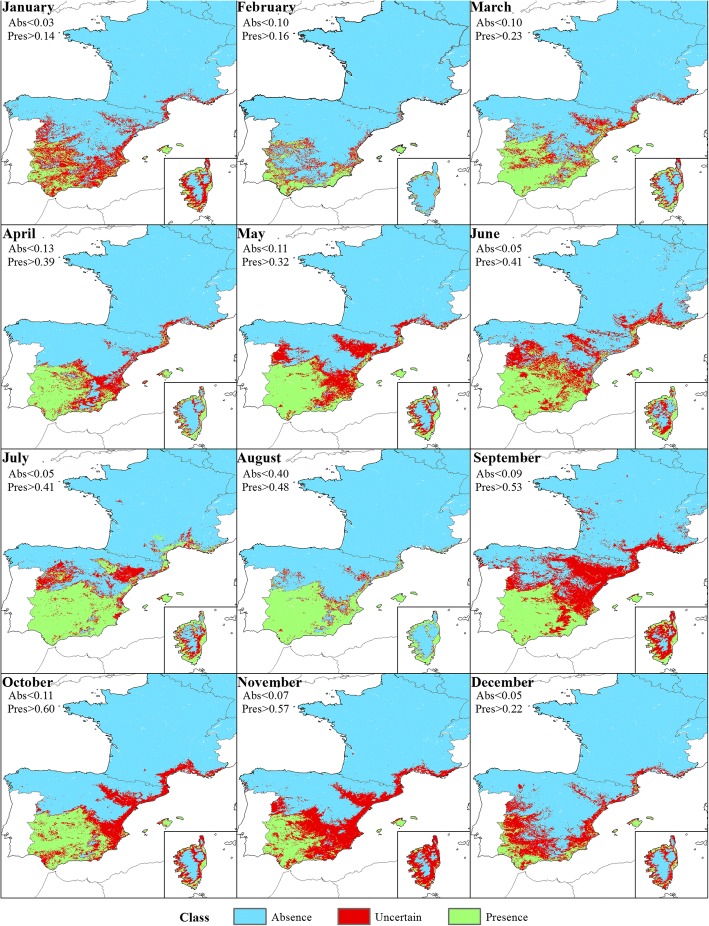
Classifications did not result in clearly delineated geographical zones for the three classes (Presence, Absence and Uncertain), although spatial patterns were observed (Fig. 4). In January, the Obsoletus ensemble was predicted present in areas within the western part of France, northern coast of Spain and in scattered areas of Germany, and it was predicted absent from northern and central Scandinavia, eastern France and parts of Germany. The Uncertain class area was present in southern Scandinavia, eastern Germany and Poland. In February the Presence area in western France and the northern coast of Spain appeared clearly segregated while more dispersed patches appeared in Germany and Poland. The Uncertain class area was reduced to patches in Germany, Poland and a small portion of southern Sweden. During March, the Presence area extended further west into France, while the Absence area was clearly concentrated in the eastern part of Europe and Scandinavia. The Uncertain area was a more coherent intermediate region between these two areas, found in eastern France, Belgium and the Netherlands. In April, the Presence class expanded from western France occupying most of France while the eastern part of the study area and Scandinavia remained in the Uncertain area. From May onwards, the general pattern showed the Obsoletus ensemble to be widely distributed in France, Germany, Austria, Switzerland, Poland and southern Scandinavia. The Absence class areas were located in southern Spain during this period. In November, Scandinavia was classified as an Absence class area together with Spain (except the northern coast of Spain, that was included in Presence area). Finally, in December the Presence class was clustered in western France and some patches in northern Germany while the remaining areas, with exception of southern Spain, appeared classified as Uncertain areas, including the Scandinavian peninsula (Fig. 4).
Fig. 4.
Classification of the predicted probability of presence of Obsoletus ensemble into Absence, Presence and Uncertain areas at a 1 km2 resolution
Pulicaris ensemble
The RF models performed less well in predicting the PP for the Pulicaris ensemble. The mean AUC was 0.81, ranging from 0.69 in April to 0.92 in December (Fig. 5).
Fig. 5.
Predicted monthly probability of presence of Pulicaris ensemble. Monthly model performance is shown as the AUC value
For January, the test set contained only three Presence observations from a single farm and the density function and thresholds could not be calculated. Therefore, the PP map could not be classified into the three classes. For February, the PP predicted for the observed Presences were completely included within the range of the PP predicted for the Absence class, meaning that the model was incapable of distinguishing the Presence class. Nevertheless, because both density functions were computed, the lower and upper thresholds were still calculated. The distribution of predicted Presence and Absence areas for the Pulicaris ensemble test set contained larger overlapping areas between both distributions than for the Obsoletus ensemble, resulting in poorer predictive power for distinguishing between the classes. For the months of April, May and June, the distribution of both classes overlapped so much that the lower threshold was calculated as close to 0 to avoid false negative classifications (Fig. 6). For the Pulicaris ensemble, the additional thresholds calculated using 10-fold CV, were similar to the main threshold for all the months, meaning that the distribution of classes in the test set were robust when subtracting 10% of the data. Both lower and upper thresholds seemed to be robust for the different test sets (Fig. 6).
Fig. 6.
Pulicaris ensemble: monthly distribution of Presence and Absence classes of the test set samples as a function of their predicted probability of presence. Dashed lines show the additional thresholds calculated from 10-fold CV
Due to the lack of Presence observations in January, we could not define thresholds for classifying the PP map. In February, because PP of the observed Presence observations were completely included in the range of the PP of the Absence class, we decided not to classify the map as the model was incapable of distinguishing the Presence class and would lead to an incorrect interpretation of the classification. In March, the Pulicaris ensemble was predicted to be present on the west coast of France, northern coast of Spain and in central and northern Scandinavia, while the Absence class was distributed in eastern France, Germany and Poland. The Uncertain area was located between the Presence and Absence class. During April, May and June, the model was able to predict the Presence class but it was incapable of distinguishing the Absence class, resulting in classification only for the Presence and Uncertain class. From July to October, the Presence class extended towards the eastern part of the study area while the Uncertain class occupied northern Scandinavia. During September, the Uncertain class was additionally found in France. In November, the Presence areas were located mostly in Germany and some patches in France while Scandinavia was classified into the Uncertain class. The Absence class was predicted in Denmark and southern Spain. During December, the Absence class was localized in Spain, France and northern Scandinavia while the Presence class remained in some patches in Germany (Fig. 7).
Fig. 7.
Classification of the predicted probability of presence of Pulicaris ensemble into Absence, Presence and Uncertain areas at a 1 km2 resolution
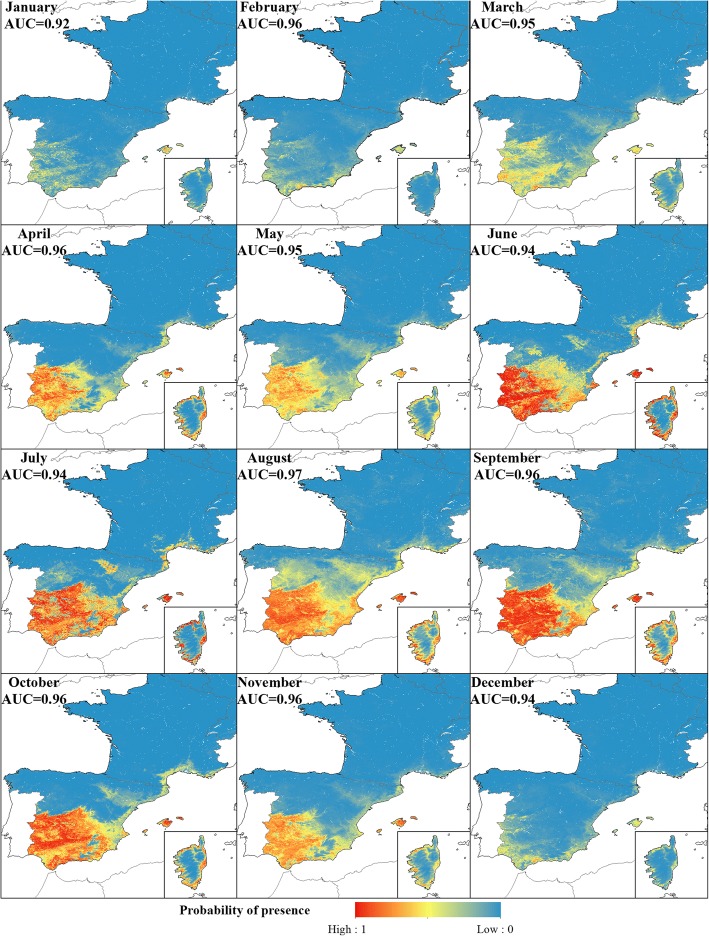
Culicoides imicola
The RF models for C. imicola had a very high accuracy for distinguishing the Presence and Absence classes. The models had a mean AUC of 0.95, ranging from 0.92 in January to 0.97 in August (Fig. 8).
Fig. 8.
Predicted monthly probability of presence of C. imicola. Monthly model performance is shown as the AUC value
The RF models predicted the C. imicola Absence class very well. Absence constituted the majority class for all months as the species was only found in Spain and southern France. The Presence class was less well predicted, as reflected in a flatter distribution. Nevertheless, the model was able to distinguish both classes, resulting in a narrow area of uncertainty between the lower and upper thresholds (Fig. 9). The additional thresholds calculated using 10-fold CV, were similar to the main threshold, indicating that the distribution of classes in the test set were robust when subtracting 10% of the data. The upper thresholds showed more variation compared to the variation in the lower thresholds. Particularly April, July and November seemed to have upper thresholds sensitive to the class distribution of the test set (Fig. 9).
Fig. 9.
Culicoides imicola: monthly distribution of Presence and Absence classes of the test set samples as a function of their predicted probability of presence. Dashed lines show the additional thresholds calculated from 10-fold CV
Compared to the models for the Obsoletus ensemble, the models for C. imicola resulted in a clearer geographical division into three separate coherent zones. Culicoides imicola was found to be present in January and February in some areas in southern Spain, the Balearic Islands and Corsica. Uncertain areas were identified in central Spain, while the Absence regions were located in northern Spain and most of France with the exception of the southern coast. From March onwards, the Presence region extended northwards, occupying the southern and central regions of Spain until October, when it retracted back to the southern coast of Spain during late autumn. On Corsica, the Presence areas were located around the coast, with the vector being absent inland. The Uncertain area was always clearly located between the Presence and Absence areas and was generally small due to the high accuracy of the model in distinguishing between Presence and Absence classes (Fig. 10).
Fig. 10.
Classification of the predicted probability of presence of C. imicola into Absence, Presence and Uncertain areas at a 1 km2 resolution
Important predictors
The most important predictors driving the distribution of the Obsoletus ensemble, Pulicaris ensemble and C. imicola were related to temperature and precipitation for most months (dLST_MN, nLST_A0, nLST_MX, BIO 10, BIO 18, BIO 5). EVI- and NDVI-derived variables were the most important for some months and for some of the taxa, but with lesser importance compared to temperature and precipitation. Corine land cover classes were not selected as important variables and only one class (CLC 12: non irrigated arable land) was selected for Pulicaris during August. A similar situation occurred for the animal density variables, in which the only variable appearing in the top 5 most important variables was sheep density for the Pulicaris ensemble. Altitude was selected as an important variable only for the Obsoletus and Pulicaris ensembles, for the month of December (Additional file 1).
Discussion
This study was based on the most extensive Culicoides dataset created to date. For these prediction maps, we used 31,429 Culicoides trap catches from nine European countries from 2007 to 2013 [33]. The objectives of this work were to predict the monthly probability of Culicoides presence and to demarcate regions of Europe into three presence classes, each for C. imicola and the Obsoletus and Pulicaris ensembles. We also identified areas and periods when the model was not able to predict with reasonable certainty. In these areas, targeted entomological surveillance programs implemented by the CVO’s of European Union member states are needed to clarify the present entomological status in case of an outbreak. The maps presented here can be used to determine vector-free areas (Absence areas) and areas where the vector can be found. The Absence and Presence areas were delimitated to minimize misclassification errors, making these classes more accurate in terms of the occurrence of Culicoides.
The models generated for the Obsoletus ensemble performed well for all months, and we were able to detect a spatial pattern in the three classes. However, the Absence and Presence classes were not completely separated by the model, and some geographical areas with Uncertain status were found among the Presence or Absence areas. For some of the months, our RF models were not able to clearly distinguish the minority class from the majority class, resulting in the threshold from the gain function being moved to the extremes to avoid misclassifications. This, in turn, resulted in a large Uncertain area that should potentially be targeted for costly entomological surveillance. This was the case for the Obsoletus ensemble during August, when the vector was indeed present in most of Europe but where our models classified the status as Uncertain in many smaller areas. For instance, in December the model predicted a large Uncertain status area that occupied most of the Scandinavia peninsula while the cold winter conditions make it unlikely that specimens will be found in northern Scandinavia. The Uncertain status areas should be interpreted with care and expert knowledge must be considered when making decisions regarding implementation of surveillance programs. The maps presented here are merely intended as tools and inputs to decision makers for long-term planning and in case of outbreaks in areas without ongoing entomological surveillance. The presented maps are based on a given gain function, but the gain function should reflect the severity of the vector borne diseases with an increasing emphasis on sensitivity as the severity of a disease increases.
In our models, the most important variables for the Obsoletus ensemble were the minimum daytime land surface temperature in January and February, and temperature- and precipitation-related variables (BIO 5 and BIO 14) throughout the rest of the year. Our results are in agreement with the findings of Calvete et al. [55] and Ducheyne et al. [56] who stated that temperature-related variables were the most important for the Obsoletus group distribution in Spain. Additionally, Purse et al [57] found that temperature had an effect in the occurrence of C. obsoletus in Italy. The Obsoletus ensemble are Palaearctic species requiring relatively low temperatures and humid climates for optimal development and survival [58, 59]. Temperature plays an important role in Culicoides ecology as it determines the seasonal fluctuation of the vector populations [60, 61], while humidity has been reported to create the optimal conditions for C. obsoletus breeding sites (e.g. dung heaps) [62].
To date, maps showing the PP and distribution of the Obsoletus ensemble for the entire Europe are scarce and incomplete. EFSA developed a website displaying distribution maps of Culicoides spp. On this site, a map of C. obsoletus/C. scoticus shows the distribution of this species [63] but the map is lacking information from some countries in Europe. At country level, some studies predicted the probability of Obsoletus group presence based on entomological data collected [56, 64–66]. Therefore, there is a need for predictions on a continental scale summarizing historical surveillance data to allow CVO’s of EU Member States to make rapid decisions in case of a future outbreak, as it would provide them with information on which areas and which time periods are likely to be vulnerable, which are likely to be safe and where the resources for surveillance should be allocated.
The RF models for the Pulicaris ensemble had poorer predictive power compared to Obsoletus ensemble and C. imicola. The abundance of the Pulicaris ensemble was ten-fold less than the abundance of the Obsoletus ensemble [33]. This lead to a lower number of Presence farms and, therefore, when the data were split into training and test sets, only a few Presence points were present in the test set. This resulted into heavily imbalanced monthly datasets e.g. February only included three farms with Presence observations in the test set. It is not recommended to assess model performance based only on a couple of observations from a certain class because it might lead to results with high variability. Culicoides pulicaris (sensu stricto) has been implicated in BTV transmission [67], but the Pulicaris ensemble species is not thought to have played a significant role in the 2006 BT outbreak in northern Europe [16]. Nevertheless, species of this ensemble might play a role in future outbreaks of emerging infections.
The model performance for C. imicola was highly accurate, with high AUC values for all months, indicating that this species has particular environmental requirements that can be detected through satellite imagery. This is likely to be related to hot and dry summers with low seasonal variation [64]: characteristic of the Mediterranean basin. The three classes were clearly distinguishable in the maps, and Presence and Uncertain areas could be delimited to the Iberian Peninsula. Culicoides imicola maps can be used directly to allocate resources for surveillance programs or to determine appropriate animal movement restrictions.
In our models, the most important explanatory variables for classification of areas for the Presence/Absence affecting C. imicola distribution were related to temperature and precipitation. We found that during winter, the mean temperature of the coldest quarter was the variable driving the presence of C. imicola, while variables related to precipitation were the most predominant drivers during the warmer months. This is in accordance with the results of previous studies [56, 64, 68].
The distribution of C. imicola has previously been modelled at continental level using classical statistical models fitted to data collected from single European countries [57, 64, 69]. In our maps, C. imicola appeared to be present all year round, as it can be found on the southern coast of Spain during January and February. This agrees with previous analyses of the start of the vector season in Europe, where C. imicola was found to be present during the winter months in southern Spain and central and southern Portugal [65]. The predicted probability of presence shown in our maps are in agreement to the distribution models made for Spain by Ducheyne et al. [56], Calvete et al. [55] and Peters et al. [70], and for France, where the Presence areas for the species are mainly located in the coastal regions of Corsica and VAR department [15].
In our study, we used Culicoides data aggregated into groups, namely the Obsoletus and Pulicaris ensembles. Aggregating species into a single group, or ensemble, might represent a challenge for ecological modelling, as the different species might require different environmental conditions and phenology differ between them. This has been studied by Searle et al. [24], who estimated the start and end date of the vector season and length of the vector-free period for four species of the Obsoletus ensemble. They observed that there were differences in phenology among the species. The lower model performance obtained for Obsoletus and Pulicaris ensembles compared to C. imicola may reflect that different species within each ensemble have different phenology and different environmental drivers. It would therefore be useful to identify Culicoides specimens to the species level. Molecular techniques, such as high-throughput real-time RT-PCR assays, can be used in a fast way for species identification. More accurate results could be expected if modelling is carried out on individual species data.
In practice, maps based on the classifications made for each 1 km2 pixel might be difficult to use for decision making, as it becomes challenging to define classes for larger areas in which pixels from different classes are found. For practical use, predicted pixel values may therefore be summarized by area, such as at NUTS level (nomenclature of territorial units for statistics) defined by Eurostat (2013). This would facilitate the implementation of control and surveillance programs by European veterinary authorities.
Random Forest is a machine learning technique that has previously been used for ecological species modelling [19, 56, 70–75]. This technique has been proven to perform better compared to other applications of classical statistical methods such as Non-Linear Discriminant Analysis and Generalized Ginear Models [19, 71], as well as Linear Discriminant Analysis, logistic regression [70, 74] and Additive Logistic Regression [75]. In this work, the monthly predicted probability of Culicoides presence had medium-high accuracy, but it is important to keep in mind that there might be other variables that cannot be captured by satellite imagery and that may have an influence on the occurrence of these species on a local scale, such as soil conditions (affecting breeding sites) and farming practices. Nevertheless, for some months, our models performed slightly better than other RF models used for predicting the occurrence of biting midges and mosquitoes [70, 71]. This highlights the challenges faced in predicting the occurrence of insect vectors using remote sensing data, as vectors are highly influenced by local microenvironments [76] and these data are difficult to obtain from satellite images without high spatial resolution.
Conclusions
We present here maps as a risk assessment tool that can be used in the future to predict potential risk areas and risk seasons for Culicoides-borne disease outbreaks. They are particularly useful for European veterinary authorities, who can classify both areas likely to have vectors and likely to be vector-free in advance and during a sudden outbreak in areas without active entomological surveillance. Predicting areas of uncertain status allows focusing costly active entomological surveillance to limited areas. The developed gain functions used to delimit the areas for targeted active surveillance can easily be adjusted to new diseases where the cost of concluding false presence or false absence may be different than suggested here.
Additional file
Table S1. The top five of the most important variables by species group for each month. The variable importance is scaled from 0 to 100. Within each month (columns), the most important variable has a value of 100. (XLSX 67 kb)
Acknowledgments
We would like to thank the Direction Générale de l’Alimentation from the French Ministry in charge of agriculture for the funding, and the Directions départementales de la protection des populations for their support in collecting the biting midges during the survey. We also thank the Ministerio de Agricultura y Pesca, Alimentación y Medio Ambiente for providing data on the national surveillance of Culicoides in Spain.
Funding
This study was funded by the EMIDA ERA-NET-supported project VICE (Vector-borne Infections: Risk-based and Cost-Effective Surveillance Systems). Culicoides data from Germany were partly collected within the German part of the VICE project funded by EMIDA ERA-NET through the Federal Office for Agriculture and Food (grant no. 314-06.01-2811ERA248). The Swiss Food Safety and Veterinary Office and the Vet-Austria project provided financial support to the Swiss and Austrian partners, respectively.
Availability of data and materials
The national surveillance and research data that support the findings of this study are available from the following people: Spain, Miguel Ángel Miranda Chueca; France, Thomas Balenghien; Germany, Jörn Gethmann; Denmark, Rene Bødker; Sweden, Anders Lindström; Norway, Petter Hopp; Poland, Magdalena Larska; Austria, Katharina Brugger; Switzerland, Alexander Mathis. Restrictions apply to the availability of these data, which were used under license for the current study and are not publicly available. Data are, however, available from the authors upon reasonable request and with permission from the national surveillance programmes of each country.
Abbreviations
- Abs
Absence class
- AUC
Area under the ROC curve
- BT
Bluetongue disease
- BTV
Bluetongue virus
- CLC
Corine Land Cover
- CV
Cross-validation
- CVO
Chief Veterinary Officer
- dLST
Daytime land surface temperature
- MIR
Mid-infrared
- NDVI
Normalized difference vegetation index
- nLST
Nighttime land surface temperature
- NUTS
Nomenclature of territorial units for statistics
- PP
Probability of presence
- Pres
Presence class
- RF
Random Forest
- ROC
Receiver operating characteristics curve
- SVFP
Seasonal vector-free period
- TFA
Temporal Fourier analysis
Authors’ contributions
ACC analysed the data and drafted the manuscript. RB planned the original study and contributed to the analysis and drafting the manuscript. LJK contributed to the analysis and drafting of the manuscript. AB and AS contributed to the analysis and interpretation of the results. HS, SAN, MGA, AL, JC, RL, SS, EK, JG, FC, ML, MS, AO, IH, SS, PH, KB, FR, TB, CG, IR, XA, JL, JCD, BM, DD, MLS, RV, BS, MAMC, CB, JL, RE, AM and WT discussed and identified preliminary national data, negotiated data access with national surveillance authorities and research projects, jointly discussed the taxonomic and spatial resolution for analysis and identified, selected and extracted the final data and relevant variables that met the criteria for the joint database. They also wrote the protocol summaries and commented on the analysis results and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ana Carolina Cuéllar, Email: anacu@vet.dtu.dk.
Lene Jung Kjær, Email: lenju@vet.dtu.dk.
Andreas Baum, Email: andba@dtu.dk.
Anders Stockmarr, Email: anst@dtu.dk.
Henrik Skovgard, Email: henrik.skovgaard@agro.au.dk.
Søren Achim Nielsen, Email: san@ruc.dk.
Mats Gunnar Andersson, Email: gunnar.andersson@sva.se.
Anders Lindström, Email: anders.lindstrom@sva.se.
Jan Chirico, Email: chirico@gmail.com.
Renke Lühken, Email: renkeluhken@gmail.com.
Sonja Steinke, Email: sonja.steinke@uni-oldenburg.de.
Ellen Kiel, Email: Ellen.Kiel@uni-oldenburg.de.
Jörn Gethmann, Email: Joern.Gethmann@fli.de.
Franz J. Conraths, Email: Franz.Conraths@fli.de
Magdalena Larska, Email: m.larska@piwet.pulawy.pl.
Marcin Smreczak, Email: smreczak@piwet.pulawy.pl.
Anna Orłowska, Email: anna.orlowska@piwet.pulawy.pl.
Inger Hamnes, Email: inger.hamnes@vetinst.no.
Ståle Sviland, Email: stale.sviland@vetinst.no.
Petter Hopp, Email: petter.hopp@vetinst.no.
Katharina Brugger, Email: Katharina.Brugger@vetmeduni.ac.at.
Franz Rubel, Email: franz.rubel@vetmeduni.ac.at.
Thomas Balenghien, Email: thomas.balenghien@cirad.fr.
Claire Garros, Email: claire.garros@cirad.fr.
Ignace Rakotoarivony, Email: ignace.rakotoarivony@cirad.fr.
Xavier Allène, Email: xavier.allene@cirad.fr.
Jonathan Lhoir, Email: jonathanlhoir@hotmail.com.
David Chavernac, Email: david.chavernac@cirad.fr.
Jean-Claude Delécolle, Email: jean-claude.delecolle@numericable.fr.
Bruno Mathieu, Email: bmathieu@unistra.fr.
Delphine Delécolle, Email: delphine.delecolle@unistra.fr.
Marie-Laure Setier-Rio, Email: mlsetierrio@eid-med.org.
Roger Venail, Email: rvenail@avia-gis.com.
Bethsabée Scheid, Email: bethsabee.scheid@ird.fr.
Miguel Ángel Miranda Chueca, Email: ma.miranda@uib.es.
Carlos Barceló, Email: carlos.barcelo@uib.es.
Javier Lucientes, Email: jlucien@unizar.es.
Rosa Estrada, Email: restrada@unizar.es.
Alexander Mathis, Email: alexander.mathis@uzh.ch.
Wesley Tack, Email: wtack@avia-gis.com.
René Bødker, Email: rebo@vet.dtu.dk.
References
- 1.Du Toit RM. The transmission of blue-tongue and horse-sickness by Culicoides. Onderstepoort J Vet Sci Anim Ind. 1944;19:7–16. [Google Scholar]
- 2.Elbers ARW, Meiswinkel R, van Weezep E. Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis. 2013;19:106–109. doi: 10.3201/eid1901.121054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellor PS, Boned J, Hamblin C, Graham S. Isolations of African horse sickness virus from vector insects made during the 1988 epizootic in Spain. Epidemiol Infect. 1990;105:447–454. doi: 10.1017/S0950268800048020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellor PS, Carpenter S, Harrup L, Baylis M, Mertens PPC. Bluetongue in Europe and the Mediterranean Basin: history of occurrence prior to 2006. Prev Vet Med. 2008;87:4–20. doi: 10.1016/j.prevetmed.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter S, Wilson A, Mellor PS. Culicoides and the emergence of bluetongue virus in northern Europe. Trends Microbiol. 2009;17:172–178. doi: 10.1016/j.tim.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Toussaint J-F, Sailleau C, Mast J, Houdart P, Czaplicki G, Demeestere L, et al. Bluetongue in Belgium, 2006. Emerg Infect Dis. 2007;13:614–616. doi: 10.3201/eid1304.061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiry E, Saegerman C, Guyot H, Kirten P, Losson B, Rollin F, et al. Bluetongue in northern Europe. Vet Rec. 2006;159:327. doi: 10.1136/vr.159.10.327. [DOI] [PubMed] [Google Scholar]
- 8.Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, Eschweiler J, et al. First occurrence of Culicoides obsoletus-transmitted bluetongue virus epidemic in central Europe. Parasitol Res. 2007;101:219–228. doi: 10.1007/s00436-007-0519-6. [DOI] [PubMed] [Google Scholar]
- 9.Zientara S, Sánchez-Vizcaíno JM. Control of bluetongue in Europe. Vet Microbiol. 2013;165:33–37. doi: 10.1016/j.vetmic.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Pinior B, Brugger K, Kofer J, Schwermer H, Stockreiter S, Loitsch A, et al. Economic comparison of the monitoring programmes for bluetongue vectors in Austria and Switzerland. Vet Rec. 2015;176:464. doi: 10.1136/vr.102979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rushton J, Lyons N. Economic impact of bluetongue: a review of the effects on production. Vet Ital. 2015;51:401–406. doi: 10.12834/VetIt.646.3183.1. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann B, Bauer B, Bauer C, Bätza HJ, Beer M, Clausen PH, et al. Monitoring of putative vectors of bluetongue virus serotype 8, Germany. Emerg Infect Dis. 2009;15:1481–1484. doi: 10.3201/eid1509.090562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S, Mcarthur C, Selby R, Ward R, Nolan DV, Mordue Luntz AJ, et al. Experimental infection studies of UK Culicoides species midges with bluetongue virus serotypes 8 and 9. Vet Rec. 2008;163:589–592. doi: 10.1136/vr.163.20.589. [DOI] [PubMed] [Google Scholar]
- 14.Dijkstra E, van der Ven IJK, Meiswinkel R, Holzel DR, van Rijn PA, Meiswinkel R. Culicoides chiopterus as a potential vector of bluetongue virus in Europe. Vet Rec. 2008;162:422. doi: 10.1136/vr.162.13.422-a. [DOI] [PubMed] [Google Scholar]
- 15.Venail R, Balenghien T, Guis H, Tran A, Setier-Rio M-L, Delécolle J-C, et al. Assessing diversity and abundance of vector populations at a national scale: example of Culicoides surveillance in France after bluetongue virus emergence. In: Mehlhorn H, et al., editors. Arthropods as Vectors. Arthropods as Vectors of Emerging Diseases. Berlin-Heidelberg: Springer; 2012. pp. 77–102. [Google Scholar]
- 16.Meiswinkel R, Baldet T, de Deken R, Takken W, Delécolle J-C, Mellor PS. The 2006 outbreak of bluetongue in northern Europe - the entomological perspective. Prev Vet Med. 2008;87:55–63. doi: 10.1016/j.prevetmed.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Hartemink N, Vanwambeke SO, Purse BV, Gilbert M, Van Dyck H. Towards a resource-based habitat approach for spatial modelling of vector-borne disease risks. Biol Rev. 2015;90:1151–1162. doi: 10.1111/brv.12149. [DOI] [PubMed] [Google Scholar]
- 18.Hay SI, Tatem AJ, Graham AJ, Goetz SJ, Rogers DJ. Global environmental data for mapping infectious disease distribution. Adv Parasitol. 2006;62:37–77. doi: 10.1016/S0065-308X(05)62002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cianci D, Hartemink N, Ibáñez-Justicia A. Modelling the potential spatial distribution of mosquito species using three different techniques. Int J Health Geogr. 2015;14:10. doi: 10.1186/s12942-015-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri S, Gilruth P, Rogers D, Szczur M. Surveillance of arthropod vector-borne infectious diseases using remote sensing techniques: a review. PLoS Pathog. 2007;3:e116. doi: 10.1371/journal.ppat.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.EFSA Panel on Animal Health and Welfare. Bluetongue: control, surveillance and safe movement of animals. EFSA J. 2017;15:e04698. [DOI] [PMC free article] [PubMed]
- 22.European Commission Ec 1266/2007. Off J Eur Union. 2007;L 283:37–52. [Google Scholar]
- 23.Brugger K, Köfer J, Rubel F. Outdoor and indoor monitoring of livestock-associated Culicoides spp. to assess vector-free periods and disease risks. BMC Vet Res. 2016;12:88. doi: 10.1186/s12917-016-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Searle KR, Barber J, Stubbins F, Labuschagne K, Carpenter S, Butler A, et al. Environmental drivers of Culicoides phenology: how important is species-specific variation when determining disease policy? PLoS One. 2014;9:e111876. doi: 10.1371/journal.pone.0111876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann C, Steinmann IC, Hegglin D, Schaffner F, Mathis A. Spatio-temporal occurrence of Culicoides biting midges in the climatic regions of Switzerland, along with large scale species identification by MALDI-TOF mass spectrometry. Parasit Vectors. 2012;5:246. doi: 10.1186/1756-3305-5-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ander M, Meiswinkel R, Chirico J. Seasonal dynamics of biting midges (Diptera: Ceratopogonidae: Culicoides), the potential vectors of bluetongue virus, in Sweden. Vet Parasitol. 2012;184:59–67. doi: 10.1016/j.vetpar.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Mehlhorn H, Walldorf V, Klimpel S, Schmahl G, Al-Quraishy S, Walldorf U, et al. Entomological survey on vectors of bluetongue virus in Northrhine-Westfalia (Germany) during 2007 and 2008. Parasitol Res. 2009;105:321–9. doi: 10.1007/s00436-009-1413-1. [DOI] [PubMed] [Google Scholar]
- 28.Clausen P-H, Stephan A, Bartsch S, Jandowsky A, Hoffmann-Köhler P, Schein E, et al. Seasonal dynamics of biting midges (Diptera: Ceratopogonidae, Culicoides spp.) on dairy farms of central Germany during the 2007/2008 epidemic of bluetongue. Parasitol Res. 2009;105:381–386. doi: 10.1007/s00436-009-1417-x. [DOI] [PubMed] [Google Scholar]
- 29.Kiel E, Liebisch G, Focke R, Liebisch A. Monitoring of Culicoides at 20 locations in northwest Germany. Parasitol Res. 2009;105:351–357. doi: 10.1007/s00436-009-1409-x. [DOI] [PubMed] [Google Scholar]
- 30.Afonso A, Abrahantes JC, Conraths F, Veldhuis A, Elbers A, Roberts H, et al. The Schmallenberg virus epidemic in Europe - 2011–2013. Prev Vet Med. 2014;116:391–403. doi: 10.1016/j.prevetmed.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann B, Scheuch M, Höper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Epizootic of ovine congenital malformations associated with Schmallenberg virus infection. Emerg Infect Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortega MD, Mellor PS, Rawlings P. Pro MJ. The seasonal and geographical distribution of Culicoides imicola, C. pulicaris group and C. obsoletus group biting midges in central and southern Spain. Arch Virol Suppl. 1998;14:85–91. doi: 10.1007/978-3-7091-6823-3_9. [DOI] [PubMed] [Google Scholar]
- 33.Cuéllar AC, Kjær LJ, Kirkeby C, Skovgard H, Nielsen SA, Stockmarr A, et al. Spatial and temporal variation in the abundance of Culicoides biting midges (Diptera: Ceratopogonidae) in nine European countries. Parasit Vectors. 2018;11:112. doi: 10.1186/s13071-018-2706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen SA, Nielsen BO, Chirico J. Monitoring of biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) on farms in Sweden during the emergence of the 2008 epidemic of bluetongue. Parasitol Res. 2010;106:1197–1203. doi: 10.1007/s00436-010-1791-4. [DOI] [PubMed] [Google Scholar]
- 35.Scharlemann JPW, Benz D, Hay SI, Purse BV, Tatem AJ, Wint GRW, et al. Global data for ecology and epidemiology: a novel algorithm for temporal fourier processing MODIS data. PLoS One. 2008;3:e1408. doi: 10.1371/journal.pone.0001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.EDENext . Biology and control of vector-borne infections in Europe. 2011. [Google Scholar]
- 37.Hijmans RJ. Free climate data for ecological modeling and GIS. 2005. Worldclim - Global Climate Data. [Google Scholar]
- 38.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 39.European Environment Agency. Corine Land Cover. 2018. https://www.eea.europa.eu/data-and-maps/data/clc-2006-raster-4. Accessed 28 Oct 2018.
- 40.Robinson TP, Wint GRW, Conchedda G, Van Boeckel TP, Ercoli V, Palamara E, et al. Mapping the global distribution of livestock. PLoS One. 2014;9:e96084. doi: 10.1371/journal.pone.0096084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Random Forests BL. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 42.Kuhn M, Johnson K. Applied Predictive Modeling. New York: Springer; 2013. [Google Scholar]
- 43.R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 44.Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:159–160. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 45.Liaw A, Wiener M. Classification and regression by randomForest. R news. 2002;2:18–22. [Google Scholar]
- 46.Breiman L. Statistical modeling: The two cultures. Stat Sci. 2001;16:199–231. doi: 10.1214/ss/1009213726. [DOI] [Google Scholar]
- 47.Guis H, Caminade C, Calvete C, Morse AP, Tran A, Baylis M. Modelling the effects of past and future climate on the risk of bluetongue emergence in Europe. J R Soc Interface. 2012;9:339–350. doi: 10.1098/rsif.2011.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce J, Ferrier S. Evaluating the predictive performance of habitat models developed using logistic regression. Ecol Modell. 2000;133:225–245. doi: 10.1016/S0304-3800(00)00322-7. [DOI] [Google Scholar]
- 49.Lunardon N, Menardi G, Torelli NROSE. A package for binary imbalanced learning. R J. 2014;6:79–89. [Google Scholar]
- 50.Batista GEAPA, Prati RC. Monard MC. A study of the behavior of several methods for balancing machine learning training data. ACM SIGKDD Explor Newsl. 2004;6:20–29. doi: 10.1145/1007730.1007735. [DOI] [Google Scholar]
- 51.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 52.Guisan A, Zimmermann NE. Predictive habitat distribution models in ecology. Ecol Modell. 2000;135:147–186. doi: 10.1016/S0304-3800(00)00354-9. [DOI] [Google Scholar]
- 53.Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Berry PM, Dawson TP, Pearson RG. Selecting thresholds of occurrence in the prediction of species distributions. Ecography. 2005;28:385–393. doi: 10.1111/j.0906-7590.2005.03957.x. [DOI] [Google Scholar]
- 55.CALVETE C, ESTRADA R, MIRANDA M. A, BORRÁS D, CALVO J. H, LUCIENTES J. Modelling the distributions and spatial coincidence of bluetongue vectors Culicoides imicola and the Culicoides obsoletus group throughout the Iberian peninsula. Medical and Veterinary Entomology. 2008;22(2):124–134. doi: 10.1111/j.1365-2915.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 56.Ducheyne E, Miranda Chueca MA, Lucientes J, Calvete C, Estrada R, Boender G, et al. Abundance modelling of invasive and indigenous Culicoides species in Spain. Geospat Health. 2013;8:241–254. doi: 10.4081/gh.2013.70. [DOI] [PubMed] [Google Scholar]
- 57.Purse BV, Tatem AJ, Caracappa S, Rogers DJ, Mellor PS, Baylis M, et al. Modelling the distributions of Culicoides bluetongue virus vectors in Sicily in relation to satellite-derived climate variables. Med Vet Entomol. 2004;18:90–101. doi: 10.1111/j.0269-283X.2004.00492.x. [DOI] [PubMed] [Google Scholar]
- 58.Purse BV, Brown HE, Harrup L, Mertens PPC, Rogers DJ. Invasion of bluetongue and other orbivirus infections into Europe: the role of biological and climatic processes. Rev Sci Tech. 2008;27:427–442. doi: 10.20506/rst.27.2.1801. [DOI] [PubMed] [Google Scholar]
- 59.Brugger K, Rubel F. Characterizing the species composition of European Culicoides vectors by means of the Köppen-Geiger climate classification. Parasit Vectors. 2013;6:333. doi: 10.1186/1756-3305-6-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purse BV, Carpenter S, Venter GJ, Bellis G, Mullens BA. Bionomics of temperate and tropical Culicoides midges: knowledge gaps and consequences for transmission of Culicoides-borne viruses. Annu Rev Entomol. 2015;60:373–392. doi: 10.1146/annurev-ento-010814-020614. [DOI] [PubMed] [Google Scholar]
- 61.Lühken R, Steinke S, Hoppe N, Kiel E. Effects of temperature and photoperiod on the development of overwintering immature Culicoides chiopterus and C. dewulfi. Vet Parasitol. 2015;214:195–199. doi: 10.1016/j.vetpar.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Steinke S, Lühken R, Balczun C, Kiel E. Emergence of Culicoides obsoletus group species from farm-associated habitats in Germany. Med Vet Entomol. 2016;30:174–184. doi: 10.1111/mve.12159. [DOI] [PubMed] [Google Scholar]
- 63.EFSA . Bluetongue virus (BTV) 2017. A story map. [Google Scholar]
- 64.Purse B, Mccormick BJJ, Mellor PS, Baylis M, Boorman JPT, Borras D, et al. Incriminating bluetongue virus vectors with climate envelope models. J Appl Ecol. 2007;44:1231–1242. doi: 10.1111/j.1365-2664.2007.01342.x. [DOI] [Google Scholar]
- 65.Ramilo DW, Nunes T, Madeira S, Boinas F, da Fonseca IP. Geographical distribution of Culicoides (Diptera: Ceratopogonidae) in mainland Portugal: presence/absence modelling of vector and potential vector species. PLoS One. 2017;12:e0180606. doi: 10.1371/journal.pone.0180606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calvete C, Estrada R, Miranda MA, Borrás D, Calvo JH, Lucientes J. Ecological correlates of bluetongue virus in Spain: predicted spatial occurrence and its relationship with the observed abundance of the potential Culicoides spp. vector. Vet J. 2009;182:235–243. doi: 10.1016/j.tvjl.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Caracappa S, Torina A, Guercio A, Vitale F, Calabrò A, Purpari G, et al. Identification of a novel bluetongue virus vector species of Culicoides in Sicily. Vet Rec. 2003;153:71–74. doi: 10.1136/vr.153.3.71. [DOI] [PubMed] [Google Scholar]
- 68.Wittmann EJ, Mellor PS, Baylis M. Using climate data to map the potential distribution of Culicoides imicola (Diptera: Ceratopogonidae) in Europe. Rev Sci Tech. 2001;20:731–740. doi: 10.20506/rst.20.3.1306. [DOI] [PubMed] [Google Scholar]
- 69.Tatem AJ, Baylis M, Mellor PS, Purse BV, Capela R, Pena I, et al. Prediction of bluetongue vector distribution in Europe and north Africa using satellite imagery. Vet Microbiol. 2003;97:13–29. doi: 10.1016/j.vetmic.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 70.Peters J, De Baets B, Van Doninck J, Calvete C, Lucientes J, De Clercq EM, et al. Absence reduction in entomological surveillance data to improve niche-based distribution models for Culicoides imicola. Prev Vet Med. 2011;100:15–28. doi: 10.1016/j.prevetmed.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 71.Ibañez-Justicia A, Cianci D. Modelling the spatial distribution of the nuisance mosquito species Anopheles plumbeus (Diptera: Culicidae) in the Netherlands. Parasit Vectors. 2015;8:258. doi: 10.1186/s13071-015-0865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ducheyne E, Charlier J, Vercruysse J, Rinaldi L, Biggeri A, Demeler J, et al. Modelling the spatial distribution of Fasciola hepatica in dairy cattle in Europe. Geospat Health. 2015;9:261–270. doi: 10.4081/gh.2015.348. [DOI] [PubMed] [Google Scholar]
- 73.Selemetas N, Ducheyne E, Phelan P, O’Kiely P, Hendrickx G, de Waal T. Spatial analysis and risk mapping of Fasciola hepatica infection in dairy herds in Ireland. Geospat Health. 2015;9:281–291. doi: 10.4081/gh.2015.350. [DOI] [PubMed] [Google Scholar]
- 74.van Doninck J, De Baets B, Peters J, Hendrickx G, Ducheyne E, Verhoest NEC. Modelling the spatial distribution of Culicoides imicola: climatic versus remote sensing data. Remote Sens. 2014;6:6604–19.
- 75.Cutler DR, Edwards TC, Beard KH, Cutler A, Hess KT, Gibson J, et al. Random Forests for classification in ecology. Ecology. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- 76.Haider N, Cuellar AC, Kjær LJ, Sørensen JH, Bødker R. Microclimatic temperatures at Danish cattle farms, 2000–2016: quantifying the temporal and spatial variation in the transmission potential of Schmallenberg virus. Parasit Vectors. 2018;11:128. doi: 10.1186/s13071-018-2709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The top five of the most important variables by species group for each month. The variable importance is scaled from 0 to 100. Within each month (columns), the most important variable has a value of 100. (XLSX 67 kb)
Data Availability Statement
The national surveillance and research data that support the findings of this study are available from the following people: Spain, Miguel Ángel Miranda Chueca; France, Thomas Balenghien; Germany, Jörn Gethmann; Denmark, Rene Bødker; Sweden, Anders Lindström; Norway, Petter Hopp; Poland, Magdalena Larska; Austria, Katharina Brugger; Switzerland, Alexander Mathis. Restrictions apply to the availability of these data, which were used under license for the current study and are not publicly available. Data are, however, available from the authors upon reasonable request and with permission from the national surveillance programmes of each country.