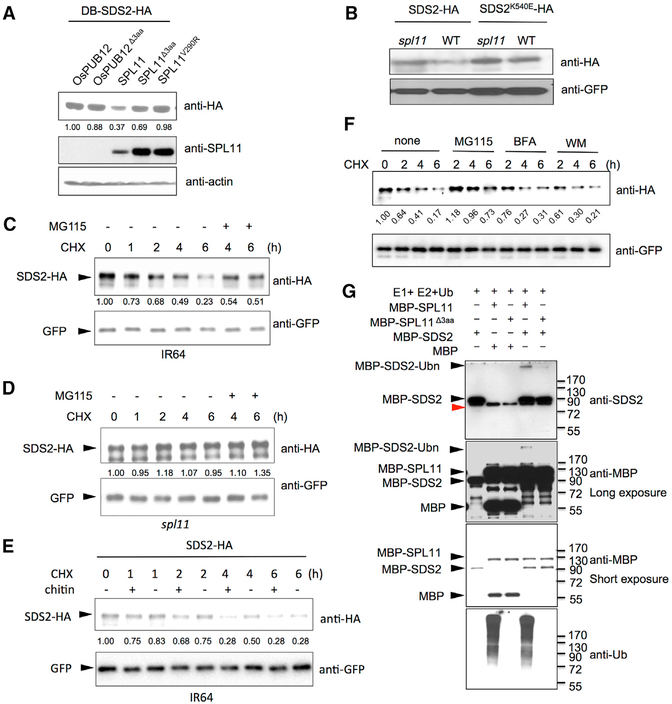
Figure 4. SDS2 Is Degraded by SPL11 and Phosphorylates the Latter.
(A) SDS2 degradation by SPL11 in yeast. SDS2-HA was co-transformed with SPL11, OsPUB12, and their mutants (SPL11Δ3aa, SPL11V290R, and OsPUB12 Δ3aa) into yeast. SDS2-HA protein level was determined by immunoblotting. Relative band intensity of each lane is labeled below the image determined by the ImageJ program.
(B) SDS2 degradation by SPL11 in rice protoplasts. Plasmids of SDS2-HA or SDS2K540E -HA was transfected into wild-type or spl11 rice protoplasts. GFP plasmids were co-transfected as a control. SDS2-HA and SDS2K540E -HA protein levels were determined by immunoblotting.
(C and D) Time course degradation of SDS2-HA in wild-type IR64 (C) and spl11 (D) rice protoplasts. Co-transfected rice protoplasts were treated with (50 μg/mL) cycloheximide (CHX) to block protein synthesis, and SDS2-HA levels were monitored by immunoblotting. 26S proteasome inhibitor MG115 (100 μM) is added to determine whether SDS2-HA is degraded via 26S Proteasome pathway after 4 and 6 hr treatment. Bands intensities determined by Image Lab software (Bio-Rad) are labeled below the bands.
(E) SDS2-HA degradation is enhanced by PAMP treatment. Transfected IR64 protoplasts were treated with 50 μg/mL CHX, 5 mg/mL chitin (+), or no chitin (−). Protoplasts are harvested at different time points after treatments and applied to western blot.
(F) Endocytosis inhibitors do not affect SDS2 degradation at the tested condition. After transfection, rice protoplasts were treated with CHX (50 mg/mL) alone or together with MG115 (100 μM), Brefeldin A (BFA, 2.0 μg/mL), or Wortmannin (WM, 0.5 μM), respectively. Protoplasts were sampled at 2, 4, and 6 hr after the treatment. Band intensities were determined by ImageJ program.
(G) In vitro ubiquitination assay of SDS2 by SPL11. MBP was used as a control substrate to show specificity. Mutant SPL11Δ3aa was used as a negative control. − and + indicate absent or present of proteins labeled on the left. Ubiquitination of MBP-SDS2 was detected by anti-SDS2 and anti-MBP antibodies. Anti-Ub antibody is applied to determine SPL11 E3 activity. Black triangles indicate corresponding protein bands labeled on the left. MBP-SDS2-Ubn denotes ubiquitinated SDS2 band.