Figure 2.
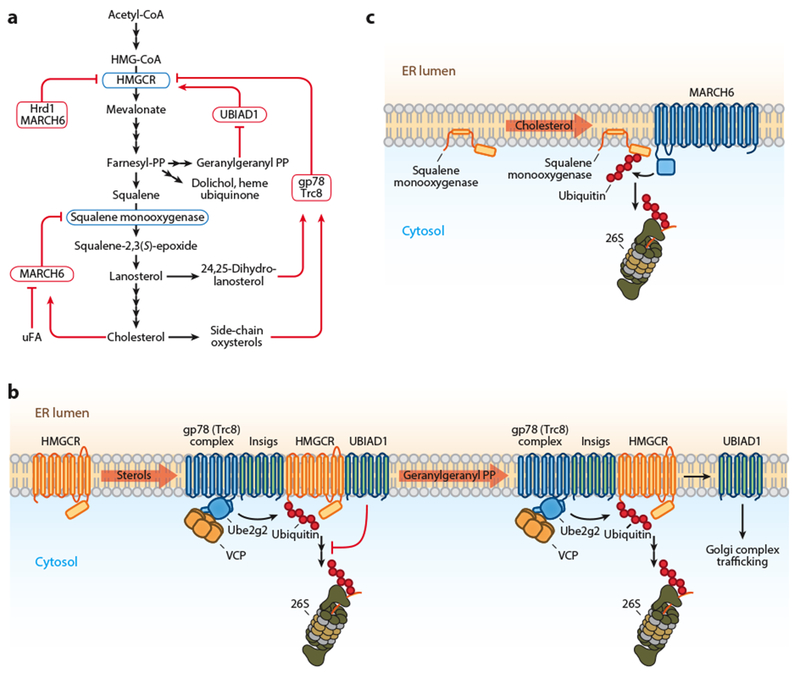
ERAD regulation of cholesterol metabolism. (a) Schematic of the cholesterol synthesis pathway with the points of ERAD-mediated regulation indicated. Regulated enzymes are shown in blue boxes and ERAD machinery in red boxes. (b) In the presence of sterols, Insigs recruit HMGCR to the gp78 and Trc8 complexes for degradation. For simplicity, only gp78 is illustrated. Sterols also stimulate the interaction of HMGCR with UBIAD1, which impairs the dislocation of a portion of HMGCR. Geranylgeranyl-PP binding releases UBIAD1 from the degradation complex, promoting HMGCR dislocation and degradation. The released UBIAD1 traffics to the Golgi complex. (c) Cholesterol induces MARCH6 degradation of squalene monooxygenase, whereas unsaturated FAs stabilize it. Abbreviations: ERAD, endoplasmic reticulum-associated degradation; FA, fatty acid; uFA, unsaturated fatty acid; gp78, glycoprotein 78; HMGCR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; Insig, insulin-induced gene protein; MARCH6, membrane-associated RING finger protein 6; PP, pyrophosphate; Trc8, translocation in renal carcinoma on chromosome 8 protein; UBIAD1, UbiA prenyltransferase domain–containing protein 1.
