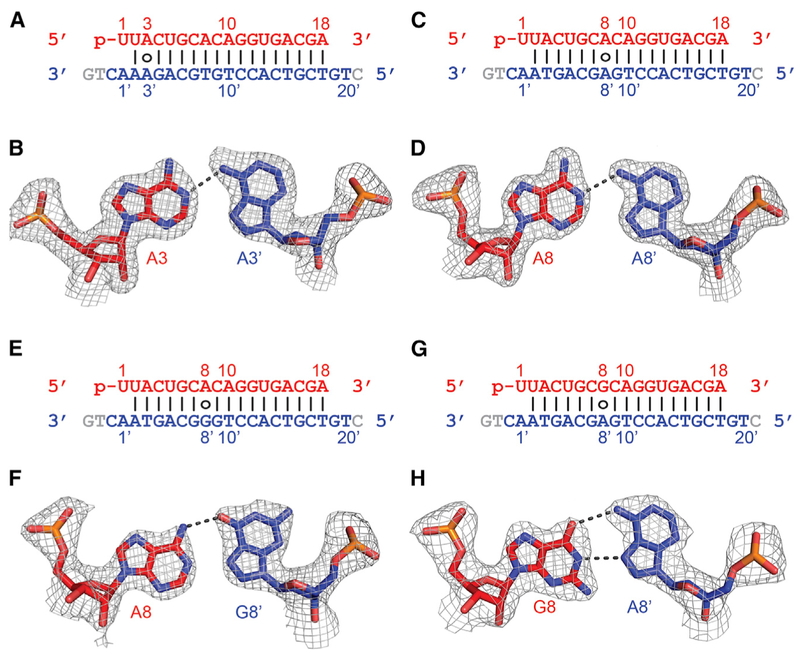
Figure 6. Pairing Alignments of A•A′, A•G′, and G•A′ Non-canonical Pairs within the Seed Segment in the Complex of RsAgo with gRNA and tDNA.
(A) Sequence alignment of the gRNA-tDNA showing the position of the A3•A3′ non-canonical pair.
(B) Pairing alignment of the cis Watson-Crick A3(anti)•Hoogsteen A3′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(C) Sequence alignment of the gRNA-tDNA showing the position of the A8•A8′ non-canonical pair.
(D) Pairing alignment of the cis Watson-Crick A8(anti)•Hoogsteen A8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(E) Sequence alignment of the gRNA-tDNA showing the position of the A8•G8′ non-canonical pair.
(F) Pairing alignment of the cis Watson-Crick A8(anti)•Hoogsteen G8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(G) Sequence alignment of the gRNA-tDNA showing the position of the G8•A8′ non-canonical pair.
(H) Pairing alignment of the cis Watson-Crick G8(anti)•Hoogsteen A8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.