Figure 4.
TMD1 of Most GPCRs Requires EMC for Optimal Insertion
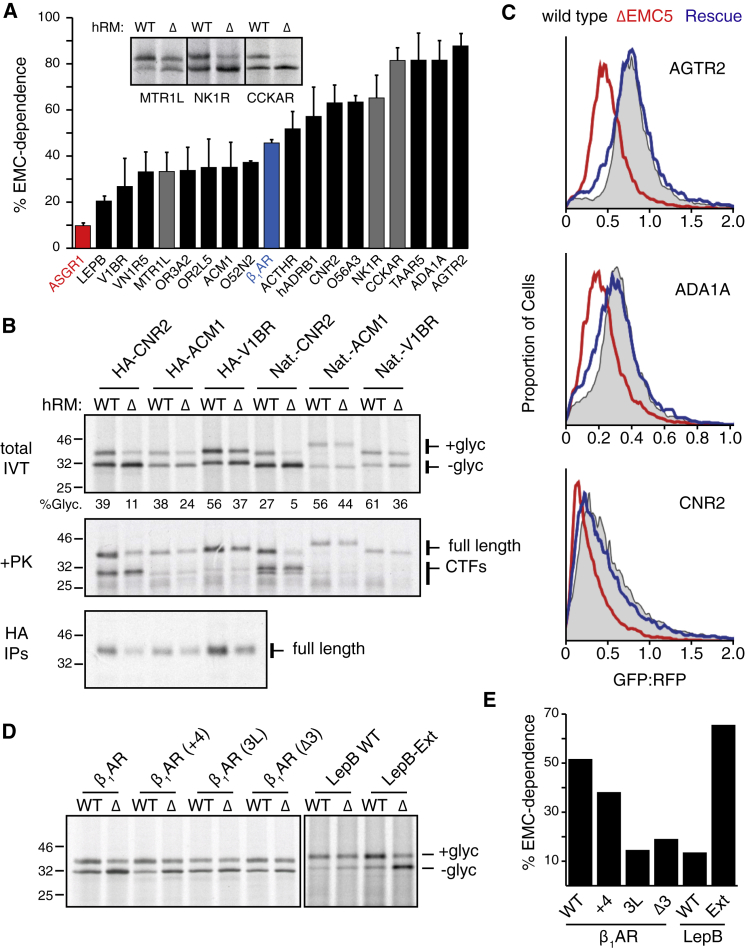
(A) Constructs containing TMD1 and flanking regions from the indicated GPCRs (see Table S1) were analyzed by glycosylation of nascent chains targeted to WT or ΔEMC6 (Δ) hRMs. The % decrease in ΔEMC6 hRM was quantified from three experiments and plotted and error bars represent standard deviation from the mean. Example data from the three GPCRs indicated by gray bars are shown in the inset. The model proteins ASGR1 and LepB were also analyzed for EMC-dependence and plotted for comparison.
(B) Ribosome-nascent chains (stalled ∼60 residues downstream of the indicated TMDs) were targeted to WT or ΔEMC6 (Δ) hRMs and analyzed by the PK-protection assay as in Figure 3C. “HA” indicates an N-terminal HA tag and glycosylation site (see Figure 3A), while “Nat.” indicates the native N-terminal domain. The PK-digested samples from the HA-containing constructs were also subjected to immunoprecipitation (HA IPs).
(C) The indicated GPCRs were tagged as in Figure 1A and analyzed by flow cytometry as in Figure 1B. Grey trace is WT cells, red trace is ΔEMC5 cells, and blue trace is EMC5-rescued ΔEMC5 cells.
(D) Ribosome-nascent chains of the indicated constructs (Table S2) were analyzed for insertion by the glycosylation assay using WT and ΔEMC6 hRMs.
(E) Quantification of the autoradiograph shown in (D).