Figure S2.
Reconstitution of EMC-Dependent β1AR Biogenesis In Vitro, Related to Figure 2
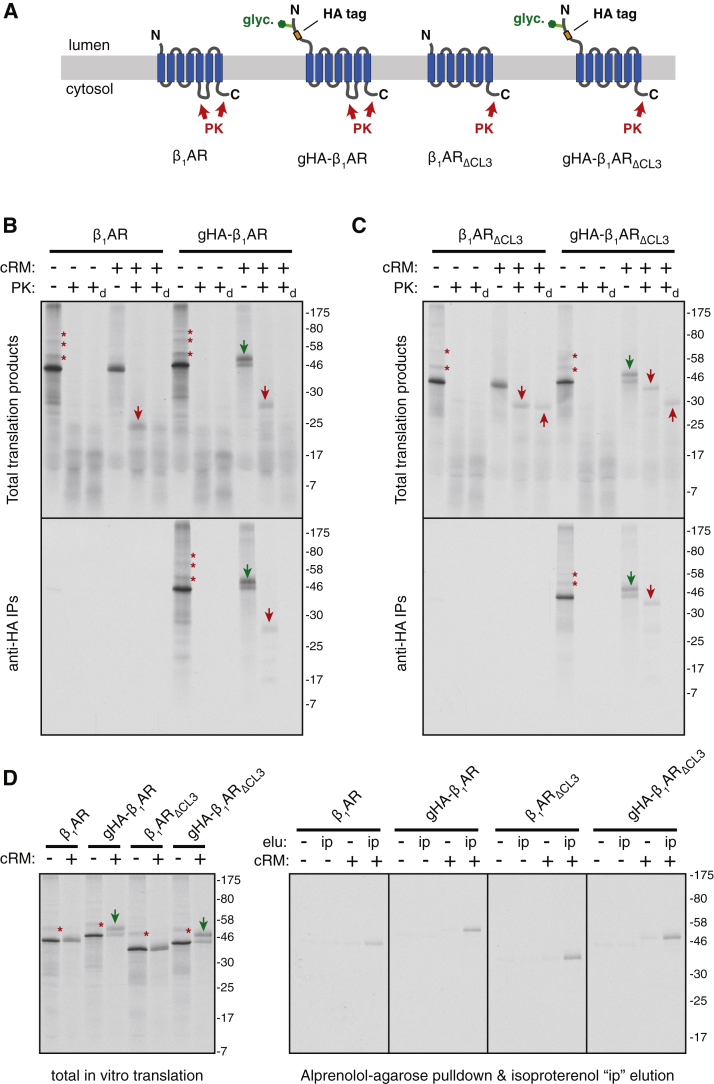
(A) Diagram of constructs used to characterize β1AR topogenesis. ΔCL3 refers to the shortening of the cytosolic loop 3 between TMD5 and TMD6. The sites that should be accessible to cytosolically added proteinase K (PK) are indicated for each construct.
(B and C) 35S-methionine labeled β1AR (or one of the indicated variants) was translated in reticulocyte lysate (RRL) in the absence or presence of canine pancreas-derived rough microsomes (cRM). The translation products were either left untreated or digested with proteinase K without or with detergent (subscripted d) as indicated. The samples were either analyzed directly (total translation products) or after immunoprecipitation via the N-terminal HA tag (anti-HA IPs) and analyzed by SDS-PAGE and autoradiography. Asterisks indicate ubiquitinated products; green arrows indicate glycosylated products; red downward arrows indicate PK-protected N-terminal fragments; red upward arrows indicate the protease-resistant 7-TMD core of β1AR left after digestion of the N- and C-terminal tails in the ΔCL3 variants. These assigned identities of the bands can be deduced by a combination of their size, change in migration upon addition of the N-terminal glycosylation site, change in digestion pattern upon shortenting of CL3 to make it proteaese-inaccessible, and IP via the HA epitope.
(D) 35S-methionine labeled β1AR (or one of the indicated variants) was translated in RRL in the absence or presence of microsomes (cRM). An aliquot of the sample was analyzed directly (total in vitro translation) or solubilized and incubated with immobilized alprenolol (a β1AR antagonist). The resin was washed, then eluted in buffer without or with isoproterenol (ip; a β1AR agonist). Efficient recovery is only observed when β1AR is synthesized with cRM and eluted with isoproterenol.