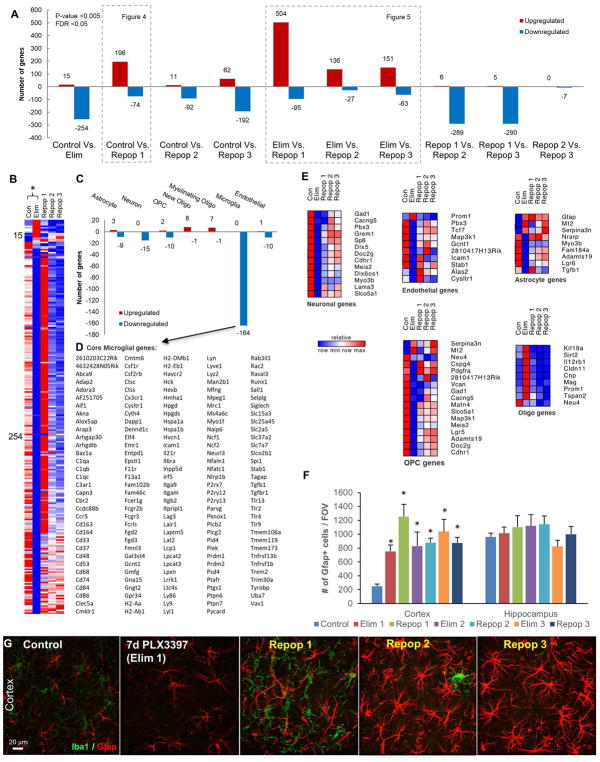
Figure 3.
Differentially expressed genes between treatment cycles with 600 ppm PLX3397, obtained via RNAseq analysis. A, Number of significant differentially expressed genes between each group analyzed. B–E, Differentially expressed genes between Con and Elim. B, Heatmap of significant differentially expressed genes. C, Differentially expressed genes stratified by those commonly expressed by CNS cell types reveals 164 downregulated microglial genes. D, List of 164 downregulated microglial genes termed “core” microglial genes. E, Heatmaps of differentially expressed genes stratified by CNS cell type reveals increase in common astrocyte-expressed genes, and decrease in common OPC-expressed genes. F–G, 63X representative images of cortex; immunostaining for Gfap and quantification of number of cells per FOV reveals the number of GFAP+ cells increases with Elim 1 and remains elevated for the remainder of cycles in the cortex, but remains unchanged in the hippocampus. All gene expression data can be accessed and searched at http://rnaseq.mind.uci.edu/green/microglia_cycles/gene_search.php. Error bars represent SEM, (n=4). * represents p<0.05 vs. Control.