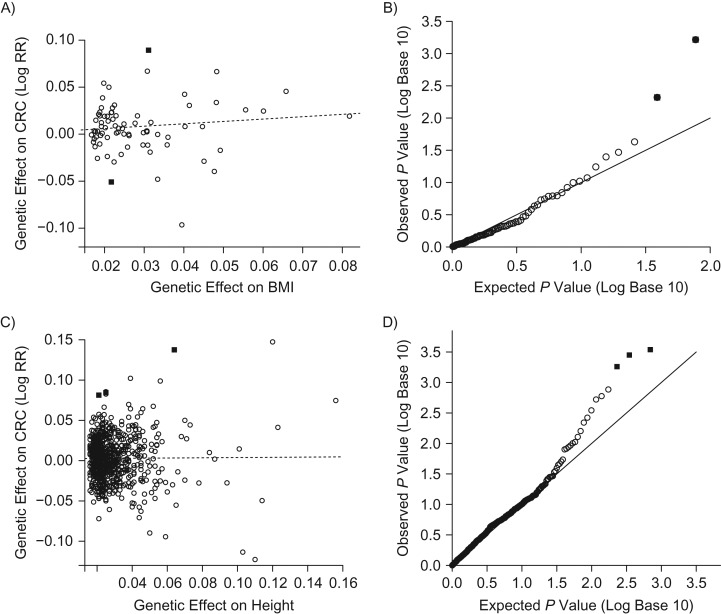
Figure 3.
Application of Menedelian randomization (MR)-Egger regression and the proposed global and individual tests for direct effects (GLIDE) test to data from the Genetics and Epidemiology of Colorectal Cancer (GECCO) Consortium in a study of body mass index (BMI; weight (kg)/height (m)2) and height in colorectal cancer (CRC) risk. The solid squares represent single nucleotide polymorphisms (SNPs) with some evidence of pleiotropy detected by GLIDE; the open circles represent SNPs which did not exhibit evidence of pleiotropy. A) MR-Egger regression results showing the intercept and slope for 77 SNPs previously shown to be linked to BMI in the Genetic Investigation of Anthropometric Traits (GIANT) Consortium (30, 31). B) Proposed GLIDE test results showing the quantile-quantile (Q-Q) plot for P values derived from assessment of surrogate direct effects for BMI. C) MR-Egger regression results showing the intercept and slope for 696 SNPs previously shown to be linked to height in the GIANT consortium. D) Proposed GLIDE test results showing the Q-Q plot for P values derived from assessment of surrogate direct effects for height. RR, relative risk.