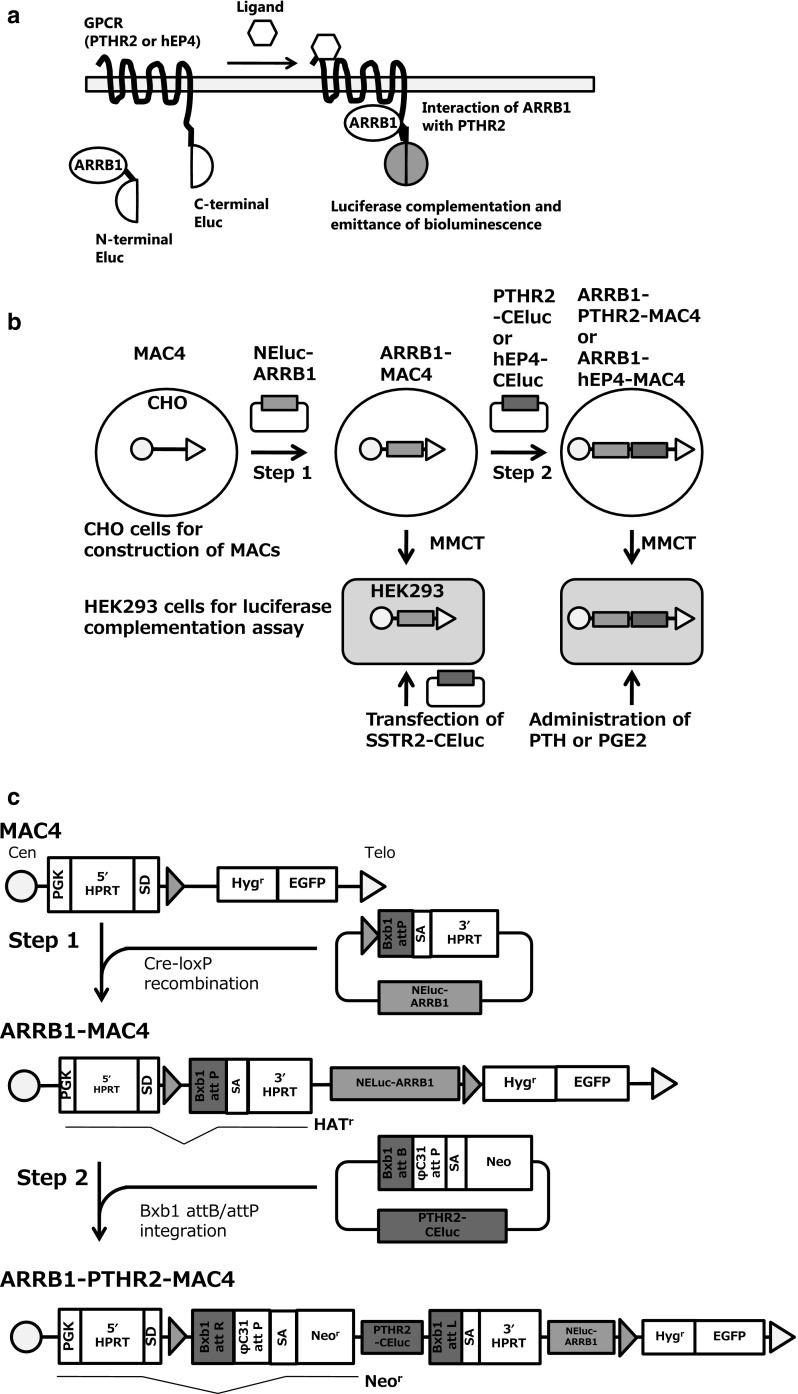
Fig. 1.
Schematic diagrams of gene loading and evaluation of luciferase activity of GPCRs using split luciferase complementation. a A schematic diagram of luciferase complementation of N-Eluc-ARRB1 and PTHR2-CEluc or hEP4-CEluc for evaluation of ligand stimulation activity. b Outline of gene loading into MACs and chromosome transfer of the MACs. ARRB1 was loaded at step 1 (ARRB1-MAC4), and then GPCRs-CEluc (PTHR2-CEluc or hEP4-CEluc) were loaded at step 2 into the MAC (ARRB1-PTHR2-MAC4 or ARRB1-hEP4-MAC4). These MACs were transferred into HEK293 cells to evaluate the bioluminescence activity by the LCA via MMCT. The SSTR2-CEluc expression vector was transiently transfected into HEK293 cells to evaluate the bioluminescence activity of ARRB1-MAC4. c Construction of ARRB1-GPCR-MAC4 using the SIM system. The SIM system enables insertion of multiple circular vectors sequentially into a MAC. It involves various kinds of site-specific recombination using Cre-loxP, Bxb1 integrase-Bxb1 attB/attP, and ϕC31 integrase-ϕC31 attB/attP. These enzymes and acceptor sites reconstitute selectable markers on the MAC. Therefore, cells acquire resistance to antibiotics when a circular plasmid is inserted into the MAC. First, NEluc-ARRB1 is inserted into MAC4 via recombination with Cre-loxP, and reconstitution of the HPRT gene leads to resistance against HAT medium (Step 1). The inserted plasmid has another specific recombination site, Bxb1 attP, which enables sequential insertion of further circular vectors with Bxb1 integrase. Second, GPCRs-CEluc, which are PTHR2-CElu or hEP4-CEluc, are inserted onto ARRB1-MAC4 via recombination of Bxb1 attB and attP with Bxb1 recombinase. The Neo resistance gene, which has a splicing acceptor site (SA) and traps PGK promoter activity instead of the HPRT gene, shows resistance against G418, and the HPRT gene is disrupted, resulting in cells with sensitivity to HAT medium. Theoretically, the φC31 attP site enables further gene loading (Step 2)