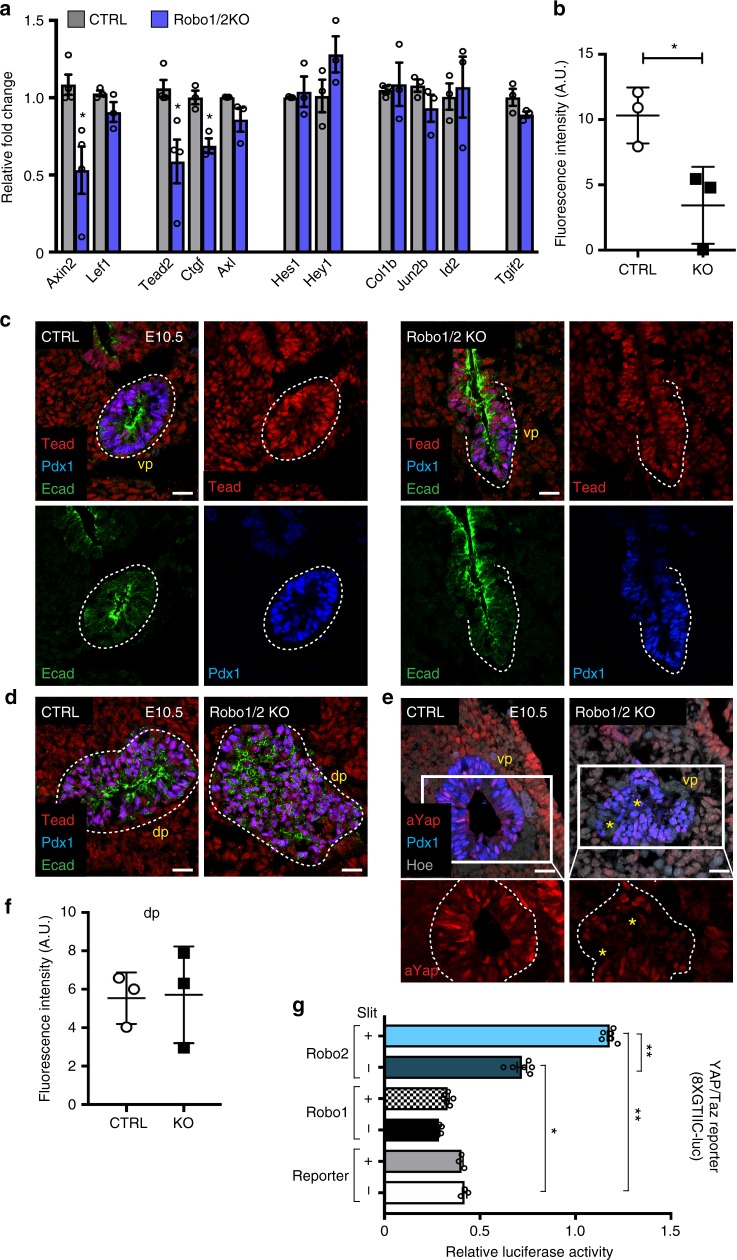
Fig. 7.
Robo signalling influences YAP/TEAD transcription activity. a RT-qPCR analysis of indicated genes in E12.5 CTRL and Robo1/2 KO mouse pancreas. Data are represented as relative fold change. Values shown are mean ± s.e.m. (n = 4). *P < 0.05, two-tailed unpaired t-test. b Quantification of panTEAD fluorescence intensity in E10.5 CTRL and Robo1/2 KO ventral pancreas normalised to Hoechst intensity and area of epithelium. (n = 3). **P < 0.01, two-tailed unpaired t-test. c Representative IF images of E10.5 CTRL and Robo1/2 KO ventral pancreas (vp) cryosections stained for Pdx1, Tead and Ecad. Merge and single channels are shown. (n = 3). d Representative IF images of E10.5 CTRL and Robo1/2 KO dorsal pancreas (dp) cryosections stained for Pdx1, Tead and Ecad. Hoechst (Hoe) was used as nuclear counterstain. (n = 3). Dashed lines mark pancreatic epithelium (c, d). Scale bars, 20 µm. e Representative IF images of E10.5 CTRL and Robo1/2 KO ventral pancreas (vp) cryosections stained for active-Yap (a-Yap), Pdx1 and Hoechst (Hoe). Insets below show boxed area at higher magnification and a-Yap channel only. Dashed lines mark pancreatic epithelium. Asterisks indicate Pdx1-low cells that are devoid of a-Yap nuclear staining. (n = 3). Scale bars, 20 µm. f Quantification of panTEAD fluorescence intensity in E10.5 CTRL and Robo1/2 KO dorsal pancreas (dp) normalised to Hoechst intensity and area of epithelium. g Dual luciferase assay in HEK293 cells transfected with the 8xGTIIC-Lux reporter and Renilla luciferase reporter plasmids, in the presence (+) or absence (−) of hRobo1-EGFP, hRobo2-EGFP, untreated or exposed to Slit recombinant proteins, as indicated. Results are expressed as the ratio of Firefly to Renilla luciferase activity. Bars are mean + s.d. **P < 0.01, *P < 0.05, two-tailed unpaired t-test. AU arbitrary units