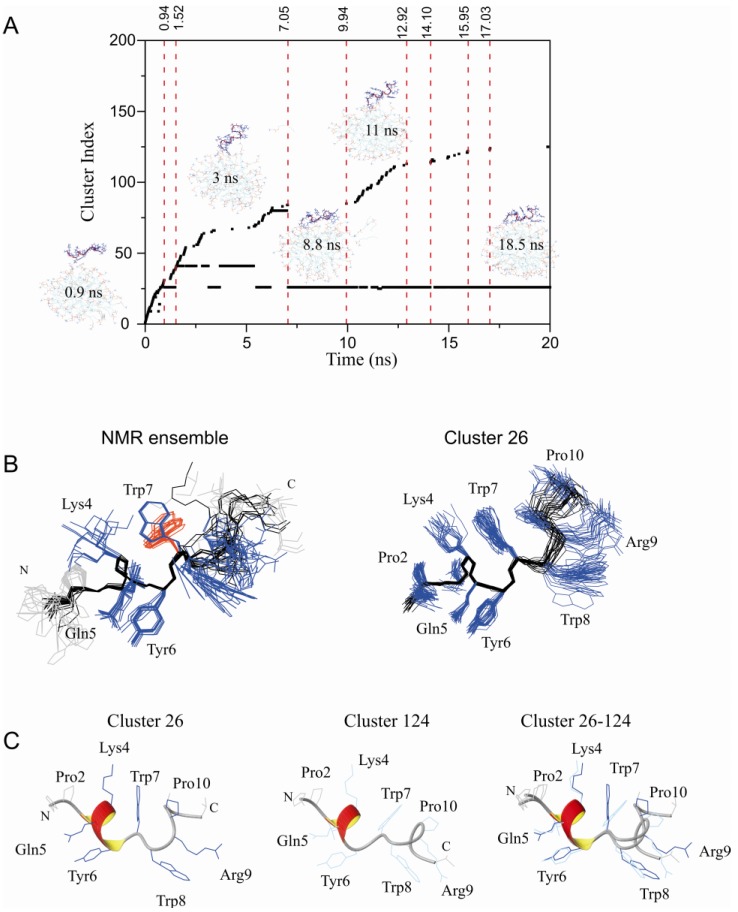
Figure 1.
Restrained MD simulation of PW2 interacting with pre-built DPC micelle. (A) Cluster index as a function of MD simulation time. Snapshots of interaction are presented at particular simulation times. In the beginning of the MD simulation, there was a rapid increase in the number of detected clusters (0 to 1.5 ns) while the peptide was approaching the membrane surface. The period after this (1.5 and 7 ns) showed binding instability where PW2 was partially bound by the N-terminal His 1 residue. After 7 ns, there was complete interaction of PW2 with the micelle surface, stabilizing cluster 26, and followed by partial dissociation events, indicated by new cluster index increments. Cluster 26 refers to the PW2 structures fully associated with the DPC micelle. (B) NMR structure ensemble (PDB ID: 2JQ2), superposition of the 20 lowest energy structures. (right) superposition of 20 representative structures of cluster 26. The backbone is in black and the side chains in blue lines. The second Trp 7 rotamer is in red lines. (C) Ribbon representation of clusters 26 (fully bound state, left) and 124 (partially dissociated state, middle) and superposition of both (right), as indicated in the figure. Side chains are represented by dark blue lines for cluster 26 and blue lines for cluster 124.